HCLS/ClinicalObservationsInteroperability/Terminology/Ontologies
representations of terminologies
Issues
Classes vs. Individuals
A given a hierarchy in some terminology:
snomed:Infectious-disease-of-lung_disorder
⇧
snomed:Infective-pneumonia_disorder
⇧
snomed:Bacterial-pneumonia_disorder
⇧
snomed:Secondary-bacterial-pneumonia_disorder
can be expressed as a class hierarchy:
snomed:Infective-pneumonia_disorder rdfs:subClassOf snomed:Infectious-disease-of-lung_disorder .
snomed:Bacterial-pneumonia_disorder rdfs:subClassOf snomed:Infective-pneumonia_disorder .
snomed:Secondary-bacterial-pneumonia_disorder rdfs:subClassOf snomed:Bacterial-pneumonia_disorder .
This is appropriate for use cases where the hierarchy is important, such as some CDS system which looks for more than one of the superclasses of snomed:Secondary-bacterial-pneumonia_disorder. If that's not required, one can use a much more efficient closure to group terms into a single-level hierarchy. For instance, if we simply want to collect all forms of infection lung diseases, we can represent the terms as a value set:
my:Infectious-disease-of-lung_disorder snomed:Infective-pneumonia_disorder owl:oneOf (
Infectious-disease-of-lung_disorder
snomed:Bacterial-pneumonia_disorder
snomed:Secondary-bacterial-pneumonia_disorder ).
This is more efficient, at the expense of expressivity (the hierarchy is gone). If we know the instance data will only use e.g. leaf-level codes, we can even eliminate the intermediate terns:
my:Infectious-disease-of-lung_disorder owl:oneOf (
snomed:Secondary-bacterial-pneumonia_disorder ). # plus viral + ...
Concept Descriptor vs. IRI
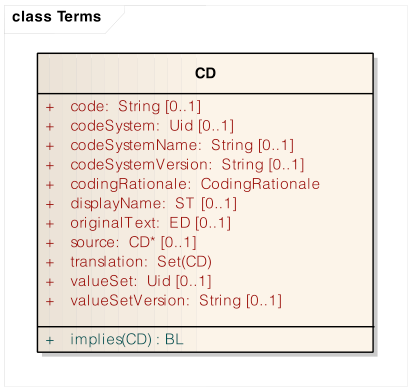
Most clinical record systems use something like an ISO 21090 CD element with attributes to uniquely identifying a concept.
The most critical these are the pair of codeSystem and code. These effectively form a unique identier for a concept. The concatonation of these can be viewed as a URN (like a URL, but not dereferencable). Using the HL7 O-RIM Coding for 21090, the attributes look like:
:labObs5678 # Joe's baseline measured in body fluid
a renal:RheumatoidFactorObservation ;
hl7:coding [ dt:CDCoding.code "13930-3" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.1" ; # or EVS or ...
dt:CDCoding.displayName
"Rheumatoid factor:Dilution Factor (Titer):Point in time:Synovial fluid (Joint fluid):Quantitative:Agglutination" ;
dt:CDCoding.codeSystemName "LOINC" ] .
Viewed as a URN, this could look like:
:labObs5678 # Joe's baseline measured in body fluid
a renal:RheumatoidFactorObservation ;
hl7:coding <urn:oidplus:2.16.840.1.113883.6.1/13930-3> .
with some helpful metadata:
<urn:oidplus:2.16.840.1.113883.6.1/13930-3>
dt:CDCoding.code "13930-3" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.1" ; # or EVS or ...
dt:CDCoding.displayName
"Rheumatoid factor:Dilution Factor (Titer):Point in time:Synovial fluid (Joint fluid):Quantitative:Agglutination" ;
dt:CDCoding.codeSystemName "LOINC" .
It is easy to use OWL equivalent class to assert the equivalence between these two forms:
<urn:oidplus:2.16.840.1.113883.6.1/13930-3>
owl:equivalentClass [
owl:intersectionOf (
[ owl:onProperty dt:CDCoding.codeSystem ; owl:hasValue "2.16.840.1.113883.6.1" ]
[ owl:onProperty dt:CDCoding.code ; owl:hasValue "13930-3" ]
) ] .
Resources
- example of LOINC as classes
- alpha prototype of LOINC in OWL format.
- LOINC Ontology (under development) captures axes.
- Another LOINC Ontology captures the complete set of LOINC codes.
- Semantic Web Representation of LOINC: an Ontological Perspective pubmed/AMIA.
- NCI CD page