HCLS/ClinicalObservationsInteroperability/FDATherapeuticAreaOntologies
Status of this Document
This is a project page for work on Semantic Web ontologies for FDA Therapeutic Areas.
Legend
This document uses some CSS styles to highlight frequently-searched text:
- design decision
- pending issue
- ericP or Charlie to study
schema or OWL model
example instance data
inline comments
Goals
- Discovery of smaller signals and more fine-grained strata.
- Query/rule re-use between trials,
- Concept re-use between Therapeutic Areas.
- Minimize cognative burden, particularly when crossing TAs.
Strategy
- Interview SMEs -- Subject Matter Experts sign off on Concept Maps presenting a graphical representation of the concepts and relationship in a TA domain.
- Model concepts in RDF/OWL.
- Re-use BRIDG where possible
- Define specific datatype and value sets.
- Re-use SIO21090 datatype definitions from O-RIM. These include complex types like uncertainty ranges, c.f. dt:TS.precision -- are they needed?
- Test models with instance data, either invented or extracted from SDTM.
- Note any place where arbitrary choices are made during synthesis -- these are points of diffusion.
- Queries performed for examination are added to ontology as endpointos.
- Specific examination criteria aren't identified in Rheumatoid Arthritis so we can't test this with data and queries.
- Use existing controlled terminoliges where possible for e.g. controlled terminologies:
- If the appropriate term is defined elsewhere, we use the existing term and its defintion. Will the SME-approved definition ever differ from that of the adopted term?
- If no appropriate term is defined elsewhere, use skos:narrowerThan links to more general terms in order to a. document terms, b. shrink the search scope for data coded with that term, c. enable the same endpoints for post-market surveillance over clinical data
Design
Modularity
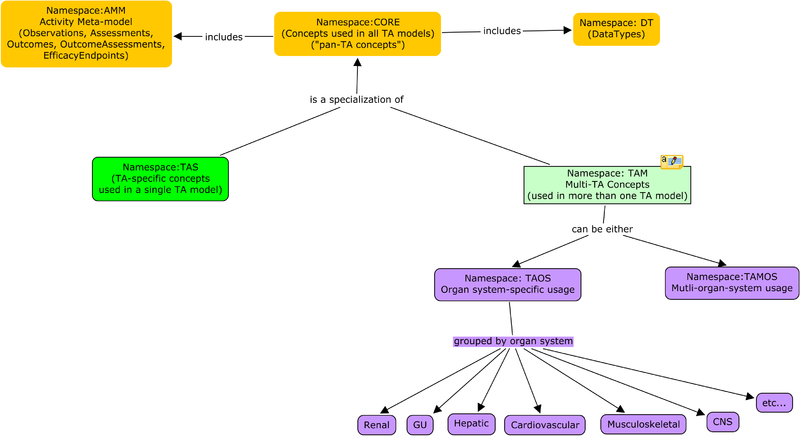
A principal goal of modeling clinical trial submissions in RDF is to leverage RDF's decentralized extensibility model based on globally-unique unambiguous identifiers for concepts and value sets. Sharing terms between submissions is a necessary component to enabling pooling of trial data. Distributed development of TAs hinges on discoverability of terms. To that end, TA-specific ontologies are broken down into:
- core ontologies -- shared datatypes, common structures like the evaluation of the effects of an intervention as a set of baseline observations, a performed intervention (e.g. substance administration), and a set of evaluation observations.
- shared ontologies -- observations, interventions and value sets shared between TA ontologies.
- TA ontologies -- capture the information required by the concept map.
| prefix | name | description | organs |
|---|---|---|---|
| integ | Integumentary | barriers, temp control | skin,hair,subcutaneous tissue |
| skel | skeletal | limb and organ support | bones, cartilage, ligaments |
| musc | Muscular | locomotion, heat production | muscles,tendons |
| cns | nervous | nervous body control and perception | brain, spinal cord, nerves, eyes, ears |
| endo | endocrine | chemical body control | pituitary, (para)thyroid, adrenal, thymus, pancreas, gonads |
| card | cardiovascular | blood transport | heart, blood, vessels |
| lymph | lymphatic | tissue fluid, immunity | speen, lymph nodes, thymus, lymphatic vessels |
| resp | respiratory | O2/CO2 exchange | lungs, trachea, larynx, nasal cavities, pharynx |
| dig | digestive | nutrient processing | stomach, GI tract, liver, pancreas, esophagus, salivary glands |
| renal | urinary | waste elimination, pH and blood volume regulation | kidneys, bladder, urethra |
| repro | reproductive | germ cell production, womb | ovaries, uterus, mammary glands testes, prostrate genitals |
For instanace, RenalTransplantation includes observations related to kidney function from renal.ttl and procedures and rejection evaluation from transplant.ttl .
Evolvability
OWL is designed to help one map ontologies where possible. One situation in which it is especially easy to map ontologies is when one has evolved from the other. This arises when changes in scientific understanding or clinical practice require changes to the model. We can break these changes down into those where the new model includes but refines the old model and those where there is a fundamental change to the data captured.
Refinement
When research reveals useful stratifications related to a therapeutic area, the related ontology can be extended to capture the new information.
If corticosteroids were found to be in two groups (perhaps by mechanism of action), the OralCorticosteroids class could be subdivided and future submissions would include that added precision.
All stratifications that worked on the original model would work on the refined data so long as the subclass inference were performed on the new subclasses of OralCorticosteroids.
New stratifications which depended on this added precision would only work on older data if that data were complemented by something which provided the precision, perhaps a drug ontology that mapped label names to the new classes.
It is also possible to change the data collected to be more general. For instance, if a topical steroids are used for rheumatoid arthritis, the appropriate ontology could have a relaxed concomitant medications list, including any corticosteroids instead of only oral corticosteroids. In this case, the route of administration would still be recorded, so any future data could be matched by conventional queries for oral corticosteroids. Future studies would permit strictly more compliant submissions, including those with topical corticosteroids.
Incompatible Change
Some changes to our world view are on "backward-compatible". For instance if clinical practice were to change from diagnosing phenylketonuria by signs of phenylalanine accumulation to using mass spectrometry, data from related studies may ...
Basis on BRIDG
In the interest of optimizing interoperability and leveraging industry interest in BRIDG, the FDA TA ontologies are mostly derived from BRIDG classes.
Domain
BRIDG is specifically limited to clinical trials. Procedures and observations are derived from from activity classes. These activity classes are scoped to stages of the clinical trial phases: definition, planning, scheduling and performance. For example, a clinical trial protocol may examine the effects of a particular drug of study which will be used in conjunction with a specific set of concomittant medications. The protocol includes prescribed administrations of the these medications as bridg:DefinedSubstanceAdministrations, usually associated with specific daytime offsets from the start of the study. A CRO will begin a study, perhaps adapting these bridg:DefinedSubstanceAdministrations to bridg:PlannedSubstanceAdministrations. When a participant enters the study, the calendar of bridg:DefinedActivities gets mapped to a calendar of bridg:ScheduledActivities. This will be populated with instances of e.g. bridg:ScheduledSubstanceAdministration. When the trial management software signals that it's time to perform some activity for a particular participant, the performed event will be associated with the defined, planned and scheduled events, for instance:
<BobAdminDrugX-1> bridg:PerformedActivity.instantiatedDefinedActivity <DrugXProtocol-Admin1> .
Property Naming
BRIDG is designed as a UML model and is expressed in RDF/OWL following the ODM 1.0 Specificaiton. This has some impact on the ontology:
- Properties are derived from bridg:attributeProperty or bridg:associationProperty.
- attributeProperties have the name of the domain in the name, e.g. bridg:DefinedObservation.targetAnatomicSiteCode.
- assocationProperties include the names of both the domain and range properties, e.g. bridg:PerformedActivity.instantiatedDefinedActivity.
- Following this naming convention means that property names are effectively single occurance over the ontology. There's no resuse of e.g. instantiated for to tie a bridg:PerformedObservation to more than one of (bridg:DefinedActivity, bridg:PlannedActivity bridg:ScheduledActivity).
- strategy: derived bridg:PerformedActivity.instantiatedDefinedActivity, bridg:PerformedActivity.instantiatedPlannedActivity, etc. from mybridg:instantiated
Structure
The majority of the data needed for efficacy analysis comes from bridg:PerformedObservationResults. BRIDG ties these back to bridg:PerformedObservations. The information in a Concept Map includes:
- observations of interest
- coding for results of these observations
- coding for consequential diagnoses and assessments
- concomittant medications
- analysis covariates
BRIDG resources
Coding specificity
BRIDG, and indeed most of clinical care, relies on shared terms identified by a tuple of a coding system identifier, a term code, and sometimes a version. The O-RIM's RDF representation of the CD.Code ISO21090 type looks like:
# Example instance data -- Sue's baseline measured in cerebral spinal fluid
:labObs1234
# Rheumatoid Factor Observations are defined in the Rheumatoid Artheritis ontology
a ra:RheumatoidFactorObservation ;
# All observations have a time
core:hasObservationTime "2013-07-07T19:02:00Z"^^xsd:dateTime ;
# We reuse the hl7 schema to capture the convention of a code and a code system (à la ISO 21090)
hl7:coding [ dt:CDCoding.code "14034-3" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.1" ;
dt:CDCoding.displayName
"Rheumatoid factor:Arbitrary Concentration:Point in time:Cerebral spinal fluid:Quantitative" ;
dt:CDCoding.codeSystemName "LOINC" ] ;
# Instance data will of course have results...
core:hasResultValue [ data:value 65 ; data:units ucum:u_mL ].
Clinical data interoperability, such as it is, relies mostly on two systems using the same code to capture an observation or procedure. The model restriction on renal:RheumatoidFactorObservation specifies that the coding have a code of some value. For a very precise restriction, the model for the above instance data looks like
# Definition of RheumatoidFactorObservation
renal:RheumatoidFactorObservation
rdfs:subClassOf
# MUST have a LOINC code of 14034-3
[ owl:onProperty hl7:coding ; owl:someValuesFrom :loinc-Rheumatoid_factor-ACnc-Pt-CSF-Qn ] .
# loinc-Rheumatoid_factor-ACnc-Pt-CSF-Qn is anything with a LOINC code of 14034-3.
:loinc-Rheumatoid_factor-ACnc-Pt-CSF-Qn
owl:equivalentClass [
owl:intersectionOf (
[ owl:onProperty dt:CDCoding.codeSystem ; owl:hasValue "2.16.840.1.113883.6.1" ]
[ owl:onProperty dt:CDCoding.code ; owl:hasValue "14034-3" ]
) ] .
# Additional subclass information relating this code to other LOINC or EVS codes:
:loinc-Rheumatoid_factor-ACnc-Pt-CSF-Qn
rdfs:subClassOf
evs:Rheumatoid_factor .
As far as I know, there are no LOINC identifiers for super classes such as a general identifier for Rheumatoid factor [EGP 2014Aug7].
The hierarchy at the bottom allows a query for a general code like evs:Rheumatoid_factor can be answered by any more instance data with a more precise code like 14034-3.
A DLQuery like RheumatoidFactorObservation and hl7:coding some <http://a.example/#evsRheumatoid_factor> will yield the instance :labObs1234.
The converse is not true is not true unless all of the more precise information is supplied in some other way (e.g. bridg:approachSite).
In order to ask specific questions of data submitted with more general codes, the submission would have to complement the codes with specifics that supplied all six LOINC axes:
- Component- what is measured, evaluated, or observed (example: urea,...)
- Kind of property- characteristics of what is measured, such as length, mass, volume, time stamp and so on
- Time aspect- interval of time over which the observation or measurement was made
- System- context or specimen type within which the observation was made (example: blood, urine,...)
- Type of scale- the scale of measure. The scale may be quantitative, ordinal, nominal or narrative
- Type of method- procedure used to make the measurement or observation
See also Mapping LOINC to SNOMED.
Use cases may motivate extending our Observation class to capture these axes (e.g. statification by site or methodology), but this would be more work than using specific codes.
The FDA TA Ontologies can:
- Insist that submitted data contain the e.g. Observation type arcs from the TA. This makes the assertion of the type arc a speach act asserting compliance with an aspect of the TA.
- Identify one or more codes, any of which imply that the Observation of of some desired type. It is sufficient, but not necessary that the data use those codes.
- Specify that the e.g. Observation use one of a set of particular code. It is necessary that the data use those codes.
For the latter two, we must pick either existing terms or invent new terms with sufficient definitions that we have "clinical compatibility" between subjects in different studies. The modeling is the same either way.
Clinical compatibility
LOINC terms are excruciatingly precise (driven by both clinical care and liability use cases) and have a consistent coded description convention for six different axes. If the submission standards are written in terms of higher-level terms like EVS Rheumatoid factor (MTHU001879), the sampled data could be radically different (e.g. kind, system, and method below), though protocol approval processes will probably limit this variability.
:labObs5678 # Joe's baseline measured in body fluid
a renal:RheumatoidFactorObservation ;
core:hasObservationTime "2012-06-06T18:01:00Z"^^xsd:dateTime ;
hl7:coding [ dt:CDCoding.code "13930-3" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.1" ; # or EVS or ...
dt:CDCoding.displayName
"Rheumatoid factor:Dilution Factor (Titer):Point in time:Synovial fluid (Joint fluid):Quantitative:Agglutination" ;
dt:CDCoding.codeSystemName "LOINC" ] ;
core:hasResultValue [ data:value 63 ; data:units ucum:u_mL ] .
An example usage shows that data taken by different lab tests in different protocols could be stratified together:
- Sue is in trial 1, Joe in trial 2.
- A reviewer is looking for signals in the aggregate of these two studies.
- Sue and Joe end up in some strata (by e.g. history and outcome).
- Resulting meta-study now has participants from both studies, with different protocol( implementation)s and potentially different e.g. measuerment sites.
It's possible that the protocol review process imposes constraints on the permissible labs (and lab codes). It would be nice for submitters, reviewers, and later analysts of aggregate data if the TA ontologies reflected at least these constraints. One outcome of this could be that multiple precise codes are permissible. This can be modelled by declaring a union of permissible values:
:RheumatoidFactorObservation
rdfs:subClassOf
# MUST have a LOINC code of 14034-3 or 13930-3
[ owl:unionOf (
[ owl:onProperty hl7:coding ; owl:someValuesFrom :loinc-Rheumatoid_factor-ACnc-Pt-CSF-Qn ]
[ owl:onProperty hl7:coding ; owl:someValuesFrom :loinc-Rheumatoid_factor-Titr-Pt-Synv_fld-Qn-Aggl ]
) ] .
:loinc-Rheumatoid_factor-ACnc-Pt-CSF-Qn
rdfs:subClassOf evs:Rheumatoid_factor ;
owl:equivalentClass [
owl:intersectionOf (
[ owl:onProperty dt:CDCoding.codeSystem ; owl:hasValue "2.16.840.1.113883.6.1" ]
[ owl:onProperty dt:CDCoding.code ; owl:hasValue "14034-3" ]
) ] .
:loinc-Rheumatoid_factor-Titr-Pt-Synv_fld-Qn-Aggl
rdfs:subClassOf evs:Rheumatoid_factor ;
owl:equivalentClass [
owl:intersectionOf (
[ owl:onProperty dt:CDCoding.codeSystem ; owl:hasValue "2.16.840.1.113883.6.1" ]
[ owl:onProperty dt:CDCoding.code ; owl:hasValue "13930-3" ]
) ] .
This is extracted from File:CodingExample.ttl .
Dereferencability
The Semantic Web (or Linked Open Data) practice of assigning dereferencable URLs assists in discovery and debugging ontologies and data.
While IHTSDO has assigned web URLs to the SNOMED ontology, retrieving them results in a Not Found error.
There is a reasonable expectation that this will be fixed in the near future and getting e.g. snomedct:77089006 will return an RDF representation of that resource.
There are, however, useful third party browsers for SNOMED which can provide useful information like http://schemes.caregraf.info/snomed#!77089006 .
These can be linked to the CD data types above (the things in the []s) with a rdfs:seeAlso.
LOINC has not assigned web URLs, however there are at least two provided by third parties. search.loinc.org provides a useful navigator which appears to have an institutional backing.
Deciderata
- What specificity provides the required clinical significance to study aggregation?
- What will inconvenience the sponsors?
- What will helpfully guide the sponsors?
- What's current practice?
- Which measurements start out with specific lab codes because those systems are in place in the care facilities?
- Which are mapped to LOINC from foreign coding systems or from no code system at all?
References
- LOINC Ontology (under development) captures axes.
- Another LOINC Ontology captures the complete set of LOINC codes.
- Semantic Web Representation of LOINC: an Ontological Perspective pubmed/AMIA.
External ontologies
The TA ontologies provide a constellation of observations and assessments associated with a particular Therapeutic Area. It's useful to connect the TA ontologies to external anatomic, physiological and disease ontologies for:
- development/governance - what observations are in other TAs which deal with inflamation of the muscular-skeletal system?
- reflection of clinical practice - does this TA ontology include the current diagnostics for a given disease?
- dependency analysis
BRIDG provides properties to connect performed observations, assessments and diagnoses to external codes.
The bridg:Activity.identifier property can identify the e.g. standard lab test identififier for an observations (however here we use the hl7:coding property in order to leverage that ontology which fully specifies ISO 21090 codes).
An observation's approach site can identify anatomical terms.
The observation result can be a bridg:PerformedObservationResult or one of its subclasses:
bridg:AdverseEventbridg:PerformedClinicalInterpretationbridg:PerformedClinicalResultbridg:PerformedDiagnosisbridg:PerformedHistopathologybridg:PerformedLesionDescriptionbridg:PerformedMedicalConditionResult
and can use the bridg:Activity.identifier to identify a pathological process or a disease.
There may be many identifiers or approach site for an observation or diagnosis:
# Example instance data -- Sue's baseline measured in cerebral spinal fluid
:labObs1234
a ra:RheumatoidFactorObservation ;
# ...
bridg:Activity.identifier
[ dt:CDCoding.code "13930-3" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.1" ;
dt:CDCoding.displayName "Rheumatoid factor:Arbitrary Concentration:Point in time:Synovial fluid:Quantitative" ; dt:CDCoding.codeSystemName "LOINC" ] ,
[ dt:CDCoding.code "54921001" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.96" ;
dt:CDCoding.displayName "Rheumatoid factor measurement" ; dt:CDCoding.codeSystemName "SNOMED" ] ,
evs:Rheumatoid_factor ;
bridg:DefinedObservation.approachAnatomicSiteCode
<http://purl.obolibrary.org/obo/DOID_676> ,
<http://umbel.org/umbel/rc/SynovialJoint> .
bridg:PerformedObservation.resultedPerformedObservationResult :labObs1234-res , :labObs1234-diag . # abbreviated as core:hasResultValue
:labObs1234-res
bridg:PerformedObservationResult.value [ data:value 65 ; data:units ucum:u_mL ].
:labObs1234-diag
a bridg:PerformedDiagnosis ;
bridg:PerformedObservationResult.identifier
doid:_676 ,
[ dt:CDCoding.code "69896004" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.96" ;
dt:CDCoding.displayName "Rheumatoid Arthritis" ; dt:CDCoding.codeSystemName "SNOMED" ] .
Note that some of these codes are identified by blank nodes with CD coding properties and some are identified directly by URL. The former is more familiar to conventional clinical practice but the latter is more convenient and appropriately leverages web technology.
Descriptions vs. Snapshots (entities)
Most clinical informatics is focused on recording detailed clinical observations rather than documenting clinical science. As an example, a LOINC code like loinc-Rheumatoid_factor-Titr-Pt-Synv_fld-Qn-Aggl above includes specifics like units and method. There are many statements one might want to make about the general concept of Rheumatoid factor, but they won't include specifics of measurement.
SNOMED addresses this by having a code for "Rheumatoid factor" which is distinct from "Rheumatoid factor measurement". SNOMED classifies the former as a substance and the latter as a procedure. We can consider LOINC and EVS codes as identifiers for procedures while the substance is appropriate for assertions associated with an ontology for clinical decision support or differential diagnostics.
SNOMED has an enormous amount of assertions reflecting physiology from the perspective of a pathologist:
snomedct:Rheumatoid_arthritis snomedct:category cnomedct:Clinical_finding ; snomedct:associated_morphology snomedct:Inflamation ; snomedct:pathological_process snomedct:Autoimmune . snomedct:Inflamation snomedct:category snomedct:Body_structure ; # seems snomedct:category little weird to me... snomedct:associated_morphology snomedct:Synovitis . snomedct:Autoimmune snomedct:category snomedct:Qualifier_value . # again, weird
and some information about tests, sometimes broken down by medium (as in LOINC's @@):
snomedct:C-reactive_protein_measurement snomedct:category snomedct:Procedure ; snomedct:code "55235003" . snomedct:Plasma_C-reactive_protein_measurement snomedct:category snomedct:Procedure ; rdfs:subClassOf snomedct:C-reactive_protein_measurement ; snomedct:code "270980008" .
See [SNOMED-CT extract] or [SNOMED-CT extract with descriptive URLs].
Capturing clinical practice
The SNOMED-CT ontology does not include assertions that capture conventional clinical practice, e.g.
snomedct:C-reactive_protein-substance ???:measures snomedct:Inflamation .
These arcs connecting observable physiological phenomena to pathologies and morphologies do not currently exist in SNOMED [EGP 2014Aug7], though it makes sense that these could come from a clinical decision support ontology. If such an ontology emerges, it can be compared against the observations and assessments in a particular TA ontologies or any of the libraries it imports. Appropriate modeling can better leverage these annotations. For instance, asserting that
snomedct:C-reactive_protein-substance ???:indicates snomedct:Rheumatoid_arthritis .
would not automatically assert that it was useful to diagnose other autoimmune reactions like lupis. Modelers will have to decide based on how general each phenomena is.
Scope
The current project is scoped to efficacy analysis, though of course much of the model incorporates data that would be needed for safety analysis. Initially, the project scope excluded protocol modeling. While the Concept Maps include the notion of concomittant medications, this property is not intrisic to the medication but instead to the use of the medication in a particular study. It's not possible to model concomittant medications without modeling protocol. There might be additional metadata in bridg:ProductRelationships.
Issues
tool choice
For
- viewing
- presenting
- developing
- mapping data to
, we choose n of:
- Protege
- TBC free
- TBC standard
- TBC maestro
.
TODO
integrate:
- -check if adequately completed- Add definitions to e.g. MH: rollup of 9b 9c 9h.
- dressing & grooming specifies a narrowerThan ("<"), but the refinement codes are on the following disabled " x " lines -- needs to reflect back to the definitions spreadsheet.
interop issues
- coding: concomittent meds and baseline observations are part of protocol
- coding: specificity of adopted codes
- coding: uncertainty ranges
- testing: do we test RA? in others, e.g. renal transplantation included "rate of X in population"
- coding: will SME defintions differ from adopted code?
- term specificity How much specificity is required for interoperability of observations between trials in a TA? Is any e.g. LOINC code for SCr good enough? Is the metric for interop whether one could transplant a patient from one study to another?
- choice between NCI EVS CUIs vs. e.g. more specific loinc and an even more specific loinc term. Using a viable submission term reduces/eliminates the requirement for a termonology server. UMLS is volatile, breaking CIUs.
- time points RenalTransplantation and ComplicatedUTI have intervals from protocol start. These are currently modeled as reusable DefinedObservations (which makes the TA sort of a template for protocols). What's the reuse utility of having two studies in the same TA have a Month3GFrObservation? What's the utility between different studies?