HCLS/ClinicalObservationsInteroperability/TermInfo
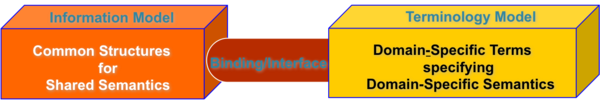
Structured clinical data is often viewed as being distributed between two "domains" (also called "views," "perspectives," "layers," "components," etc.) For the purposes of this discussion, we refer to those two "component parts of a fully-formed semantic utterance of clinical data" through by visualizing them as being distributed between two models: an Information Model and a Terminology Model. Note that the term "model" is used rather specifically in the context of this discussion for the purposes of building a concrete example, the terms Information Model and Terminology Model refer to specific, well-defined model, i.e. the HL7 RIM and the SNOMED-CT models. However the concepts of Information Model and Terminology Model in the larger discussion of the full representation of a fully-formed semantic utterance need not be restricted to a single Information Model or single Terminology Model. The critical feature about the separation of the two components is that each contains its own semantics, i.e. its own concepts, relationships, etc. In general, the Information Model expresses a network of generic concepts such roles, actions, observations and their inter-relationships, etc. -- this collection of information (small"i") is also often referred to as "meta-data" or "contextual knowledge." In contrast, the information (small "i") represented by the concepts, relationships, etc. in theTerminology Model provides specific "instance-level" descriptions of higher-order constructs. It is often heavily reliant on "is-a" and "a-part-of" relationships between concepts, and is sometimes referred to as the "data layer" or "definitional knowledge model." This is the conceptual view of the world, a view in which the Information and Terminology Models provide complementary semantics and therefore at least implicitly are clearly, cleanly, and consistently related to each other via some sort of "binding interface.
Conceptual View of the relationship between Information and Terminology models (hover over graphic for legend).
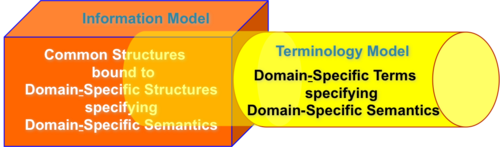
Operationally, however, the semantic expressiveness of the two models often overlaps, i.e. a given semantic statement can often be partially (or even completely) represented using a number of different combinations of concepts from the Information and Terminology models with each representation selecting some concepts and constructs from an Information Model and others from a Terminology Model. Unfortunately, this overlap in the semantic expressiveness of the two models enables multiple organizations tasked with representing the same semantic utterance to produce representations that, when serialized for transport between systems (for example, to XML) are not interoperable (hover over graphic for legend).
Example: A clinical diagnosis based on multiple observations and relationships
The following clinical diagnosis
Grade 4 anaphylactic reaction to penicillin as evidenced by the combination of wheals (hives), acute respiratory distress (ARD) and systemic hypotension (LBP) following a penicillin injection
can be represented in multiple ways depending on what semantic aspects of the overall statement one chooses to represent using a specific Information Model and its associated concepts and constructs vs the representation of the same semantics using concepts and constructs drawn from a particular Terminology Model. If done correctly, i.e. if the same "amount" of semantic content is <<explicitly>> represented in more than one representational scheme, the various representations would be viewed as clinical identical in terms of the information that they convey. Following the act of representing the semantic statement (the "design-time" form of the statement), each resulting complex construct must be serialiazed into some form of "byte stream" (usually XML, as will be used for the purposes of this discussion) for transmission between systems (the "run-time" form of the statement). Unfortunately, different design-time representations result in different run-time serializations with the high likelihood that two desing-time representations known to be semantically equivalent by human authors or reviewers will be analyzed to be <<computationally>> non-interoperable because the order of elements in XML is critical to automated determination of semantic equivalence.
The following example uses the HL7 Reference Information Model and the SNOMED-CT Terminology Model to demonstrate both the problem -- i.e. different representations serialized using HL7's standard RIM-to-XML serialization protocol ("HL7 XML Implementation Technology Specification (XML ITS)) are not computationally interoperable from the perspective of semantic equivalence -- as well as to propose a solution using Semantic Web tools and technologies. Specifically, in the Unification section, we show how multiple design-time representations -- when expressed in RDF permit in the presence of pre-existing RDF representations of bot the RIM and SNOMED -- allow one to computationally determine semantic equivalence in spite of differences in run-time serializations that would otherwise be analyzed by machines as being non-interoperable, i.e. would not be recognized as semantically equivalent if standard XML parsing strategies were applied.
Semantics primarily represented in Information Model
Atomized RIM XML
The RIM/XML for the example diagnosis can distribute individual SNOMED codes across multiple, finely-graunulated RIM Observation class instances. These instances are, in turn, semantically linked using Source ("inbound") and Target ("outbound") ActRelationships typeCode values. (NOTE: the semantics of the various tyepCode relationships are predefined in a vocabulary published by HL7 and controlled through the HL7 RIM Harmonization process to prevent semantic redundancy or "one-off" creation of rogue ActRelationship.typeCode values.)
<content xsi:type="Observation" classCode="OBS" moodCode="EVN">
...<value code="241938005" displayName="penicillin-induced anaphylaxis (disorder)" .../>
<inboundRelationship typeCode="SUBJ">
<source ...>
<code code="SEV" displayName="Severity Observation" .../><value code="423132009" .../>
</source>
</inboundRelationship>
<outboundRelationship typeCode="EVID">...
<code code="ASSERTION" .../><value code="247472004" displayName="weal (disorder)" .../>...
</outboundRelationship>
... <!-- etc. for 373895009 (acute respiratory distress) and 45007003 (low blood pressure) -->
</content>
Atomized RIM RDF
Expressed as RDF (turtle), this diagnosis captures the ActRelationships from the information model, but the semantics of the terminology are still just numeric literals: @@ captures weal, ARD, LBP, severity but not Substance Administration Act @@
# penicillin-induced anaphylaxis (disorder) :
_:diagnosis5 a rim:Observation ;
rim:Observation.value [ dt:CD.code "241938005" ];
# severity = 423132009 | grade 4 out of 5 |
[ a rim:ActRelationship ; ...
rim:ActRelationship.code [ dt:CD.code "rim:SEVERITY" ] ;
rim:ActRelationship.source _:diagnosis5 ;
rim:ActRelationship.target _:severity ] .
_:severity a rim:Observation ;
rim:Observation .value [ dt:CD.code "423132009" ] .
# has definitional manifestation = 247472004 | weal (disorder) |
[ a rim:ActRelationship ; ...
rim:ActRelationship.code [ dt:CD.code "rim:ASSERTION" ] ;
rim:ActRelationship.source _:diagnosis5 ;
rim:ActRelationship.target _:weals ] .
_:weals a rim:Observation ;
rim:Observation .value [ dt:CD.code "247472004" ] .
# etc. for 373895009 (acute respiratory distress) and 45007003 (low blood pressure)
or in a simplified (non-reified) representation:
# penicillin-induced anaphylaxis (disorder) :
_:diagnosis5 a rim:Observation ;
rim:Observation.value [ dt:CD.code "241938005" ];
rim:SEVERITY [ rim:Observation.value [ dt:CD.code "423132009" ] ;
rim:ASSERTION [ rim:Observation.value [ dt:CD.code "247472004" ] .
# etc. for 373895009 (acute respiratory distress) and 45007003 (low blood pressure)
Semantics primarily represented in Terminology Model
This same diagnosis can be expressed as a single Observation in the information model with a complex, post-coordinated SNOMED code. This SNOMED code fully specifies the semantics of the entire clinical statement and can thus be bound to a single instance of a RIM Observation class:
241938005 | penicillin-induced anaphylaxis (disorder) | : 246112005 | severity | = 423132009 | grade 4 out of 5 | , 363705008 | has definitional manifestation | = 247472004 | weal (disorder) | , 363705008 | has definitional manifestation | = 373895009 | acute respiratory distress | , 363705008 | has definitional manifestation | = 45007003 | low blood pressure (disorder) |
Terms like 246112005 | severity | and 363705008 | has definitional manifestation | correspond to types of ActRelationships in HL7 RIM, in particular SUBJ and EVID. See the Mapping from SNOMED attributes to V3
Collected RIM RDF
# penicillin-induced anaphylaxis (disorder) :
_:dx ...
rim:Observation.value [ dt:CD.code
"241938005:246112005=423132009,363705008=247472004,..." ].
This representation represents a "transfer" of much of semantic expressiveness from the Information Model (RIM) to the Terminology Model (SNOMED-CT). This transfer is evidenced by the marked <<decrease>> in the number of RIM instances required along with the concomitant <<increase>> in the complexity of the SNOMED code. From a clinical perspective, both representations are semantically equivalent.
It is important to note that two "design-time" representations developed above --the "poles" of different approaches -- are but two of a fairly large number of semantically equivalent but representationally different constructs that could be developed using the RIM and SNOMED-CT. An intermediate representation between the two poles is presented below to emphasize the heterogenous and ultimately somewhat stylized nature of the various representations. The important point is that each design-time representation is semantically equivalent, but that because each representation would result in a different "run-time" XML serialization, automated processing of the various run-time artifacts would often determine them to be non-interoperable, i.e. would not recognize their semantic equivalence.
Intermediate representations
The completely Atomized and completely Collected representations of the diagnosis represent two poles of a continuum of possible expressions. A representation with multiple Observations for weal, ARD, LBP but additional Observations for Grade IV anaphylaxis, single Substance Administration Act is a reasonable example between these poles:
Intermediate RIM RDF
# penicillin-induced anaphylaxis (disorder) :
_:dx ...
rim:Observation.value [ dt:CD.code "241938005:246112005=423132009" ];
rim:Act.outboundRelationship _:weals, _:respDistress, _:lbp .
# has definitional manifestation = 247472004 | weal (disorder) |
_:weals a rim:ActRelationship ; ...
rim:ActRelationship.target [ rim:Observation.value [ dt:CD.code "247472004" ] ] .
# etc. for 373895009 (acute respiratory distress) and 45007003 (low blood pressure)
Unifying these representations
We can Add an extra property to each Observation which captures the semantics of the potentially complex terminology code. In the case of SNOMED's compositional terms, the semantics can vary from simply recapitulating a soul number to creating complex objects with their own properties and attributes. Mechanically, this involves micro-parsing the dt:CD.code attributes and attaching them to the Observations via a new terminfo:termcode arc. Applying this to the Collected representation, we see a termcode which may be reasoned about with OWL or SPARQL:
Annotated collected RIM RDF
# penicillin-induced anaphylaxis (disorder) :
_:dx ...
rim:Observation.value [ dt:CD.code
"241938005:246112005=423132009,363705008=247472004,..." ];
terminfo:termcode [
a <http://www.ihtsdo.org/SCT_241938005> ;
<http://www.ihtsdo.org/SCT_246112005> <http://www.ihtsdo.org/SCT_423132009> ;
<http://www.ihtsdo.org/SCT_363705008> <http://www.ihtsdo.org/SCT_247472004>
# etc. for 363705008=373895009 and 363705008=45007003
];
Alternative
# penicillin-induced anaphylaxis (disorder) :
@prefix sctid: <http://www.ihtsdo.org/SCT_> .
_:dx ...
rim:Observation.value dt:CD.code [
skos:notation "241938005:246112005=423132009,363705008=247472004,...";
a sctid:241938005 ;
sctid:246112005 sctid:423132009 ;
sctid:363705008 sctid:247472004
# etc. for 363705008=373895009 and 363705008=45007003
] .
We can now write OWL or SPARQL rules to examine the predicates associated with the 12 axes of SNOMED (e.g. SCT_246112005, SCT_363705008) to map them into their equivalent expressions in the information model.
# penicillin-induced anaphylaxis (disorder) :
CONSTRUCT {
?diag rim:Act.outboundRelationship [
a rim:ActRelationship ; ...
rim:ActRelationship.target [ rim:Observation.value [ dt:CD.code "?code" ] ]
} WHERE {
?diag rim:Act.outboundRelationship [
terminfo:termcode [
<http://www.ihtsdo.org/SCT_363705008> ?codeURL
]
]
BIND (substr(?codeURL, 26) AS ?code)
}
Reasoning with the terminology model
SNOMED codes have a normalized form; the normalized form of the above SNOMED is:
39579001 | anaphylaxis | :
246075003 | causative agent | = 373270004 | penicillin -class of antibiotic- | ,
246112005 | severity | = 423132009 | grade 4 out of 5 |,
263502005 | clinical course | = 424124008 | sudden onset AND/OR short duration | ,
363705008 | has definitional manifestation | = ( 247472004 | weal |:
{ 116676008 | associated morphology | = 1806006 | eruption | ,
363698007 | finding site | = 39937001 | skin structure | }),
363705008 | has definitional manifestation | = ( 373895009 | acute respiratory distress | :
363698007 | finding site | = 20139000 | structure of respiratory system | ,
363714003 | interprets | = 248546008 | ease of respiration | ,
363714003 | interprets | = 278844005 | general clinical state | ) ,
363705008 | has definitional manifestation | = ( 45007003 | low blood pressure | :
363698007 | finding site | = 281159003 | systemic arterial structure |
)
Having the SNOMED terms in the same domain of discourse as the HL7 V3 allows us to use OWL for SNOMED gives RDF tooling access to this and even more rich information. Following are some excerpts from the relevant OWL axioms:
"Micropapular weak is a disorder __located in__ the skin, and a __morphology__ of Maculopapular rash"
<owl:Class rdf:about="SCT_298138003">
<rdfs:label xml:lang="en">Micropapular weal (disorder)</rdfs:label>
<rdfs:subClassOf><owl:Class>
<owl:intersectionOf rdf:parseType="Collection">
<owl:Class rdf:about="SCT_247472004"/>
<owl:Restriction>
<owl:onProperty rdf:resource="RoleGroup"/>
<owl:someValuesFrom>
<owl:Class>
<owl:intersectionOf rdf:parseType="Collection">
<owl:Restriction>
<owl:onProperty rdf:resource="SCT_363698007"/>
<owl:someValuesFrom rdf:resource="SCT_39937001"/>
</owl:Restriction>
<owl:Restriction>
<owl:onProperty rdf:resource="SCT_116676008"/>
<owl:someValuesFrom rdf:resource="SCT_47725002"/>
</owl:Restriction>
</owl:intersectionOf>
</owl:Class>
</owl:someValuesFrom>
</owl:Restriction>
</owl:intersectionOf>
</owl:Class></rdfs:subClassOf>
</owl:Class>
These axioms allow us to query richer representations of and/or relationships to the original diagnostic statement, e.g. associations between drug administration and other reparatory or cardiovascular symptoms.