HCLS/ChemicalTaxonomiesUseCase
Chemical Taxonomy and Structure Use-Cases
Annotating and classifying chemical compounds using the semantics popularly accepted by researchers involved in drug design is crucial for the effective use of Biological and structural data for technological growth and drug development. Drug design is a collective and coordinated effort involving several disciplines such as structural biology and medicinal chemistry. The semantics, chemical taxonomies, used by these disciplines may differ from one another. Several semantic representations of the views commonly used by these disciplines are presented and their utilization in developing a chemical Semantic Web is illustrated.
Further reading, downloads, and publications are available at: http://xpdb.nist.gov/hiv2_d/download.html. For further information, contact T.N. Bhat (bhat@nist.gov) or John Barkley (jbarkley@nist.gov). Inspiration and guidance in the preparation of this use case by Eric Neumann, Anne Plant, & Henry Rodriguez is greatly appreciated.
3D structures of HIV protease inhibitors
Structural Biology and Modeling View
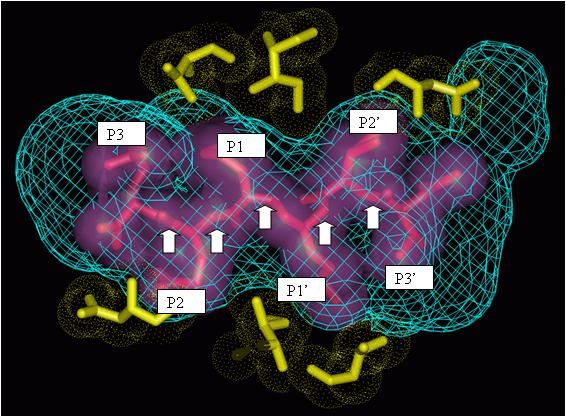
A structural biologist facilitates drug design by studying enzyme drug interactions using a computer generated model built using experimental X-ray or NMR data. The popular paradigm in drug discovery seeks to collect, compare, and test many chemically similar compounds built by linking several building blocks generally known as fragments. This approach to modern drug discovery[1] is rational and knowledge-based with a defined hypothesis on the functional role of individual fragments that make up a drug. The process of drug design, therefore, begins with a lead compound followed by a hypothesis as how different fragments of this compound interact with the amino acid residues of the target protein molecule[2]. Following this, database searches are performed to gather structural neighbors of the lead compounds using what is commonly known as ‘mix and match method of building the functional fragments’ of a lead compound. The suitability of these fragments that makeup a lead compound is established using fragment-based in silico modeling or X-ray crystallographic screening techniques[3,4] These fragments are reviewed based on complimentarity between the properties of the fragments and the enzyme residues. Charge – charge complimentarity (pairing of + & - charges), hydrogen-bond complimentarity (donar & acceptor pairing), space filling complimentarity (void and volume pairing) are a few of these properties.
The above figure shows interactions between the amino acid residues of HIV protease (gold) and drug fragments (pink). Modelers usually divide drugs into fragments such as P1, P2, P3, P1’, P2’, and P3’ by breaking the drugs at the peptidic bonds (marked by white arrows). Often these fragments are further classified based on the type of the atoms and/or the type of the ring structures they have. Additional such pictures may be viewed at http://xpdb.nist.gov/hivsdb/gallery.html or here.
Alarmed at the sharply rising cost of carrying out such experiments involving thousands of chemicals, researchers are more and more looking for help from Bioinformatics and Information Technology. Chemical databases play a major role in holding, annotating and distributing chemical, biological, medicinal, and structural data (e.g., HIVSDB http://xpdb.nist.gov/hivsdb/hivsdb.html, Enhanced NCI Database Browser - http://cactus.nci.nih.gov/; Chemistry WebBook -http://webbook.nist.gov/chemistry/; Thermodynamics Research Center - http://www.trc.nist.gov; Mass Spectral Library - http://www.nist.gov/srd/; Protein Data Bank - http://www.rcsb.org/pdb/Welcome.do; Relibase - http://relibase.ebi.ac.uk/reli-cgi/rll?/reli-cgi/general_layout.pl?home). Such Web sites are an important part of the current means of information exchange on chemical structures (compounds). However, these databases do not interoperate. To date a fully mature example of a Chemical Semantic Web is lacking. We illustrate a use case for some of the Semantics of a Chemical Web using chemical structural data for AIDS. This use case can also take advantage of interactions between compounds and protein targets, as is defined in the Small Molecules Interaction Database (SMID) that is described in the Small Molecules that Bind to Proteins Use-Case.
Chemical structures are complicated because of the many ways a given set of atoms may come together in a three-dimensional space to form compounds (drugs). The most effective way of representing chemical structures is by their schematic drawings that show details of their mutual disposition using chemical scaffolds, such as, ring structures, type of bonds, type of atoms and hybridization. Here we illustrate the use of schematic drawings in combination with IUPAC names and InChI[5] to define Semantic concepts, statements and conjugated statements to formulate some of the popular questions raised during a drug design process.
Elementary Questions Asked by the Structural Biologist and Modeler
What fragments are available for designing a new drug?
Concepts – chemical, fragment. A chemical or drug has ‘one to many relationship to a fragment. A chemical may have one or more fragments; a given fragment may be common among one or more chemicals.
Examples of fragments:
- IUPAC Name: 2-isopropyl-1,3-thiazole
Schematic Drawing: 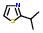
Statement: 2-isopropyl-1,3-thiazole is-a fragment or  is-a fragment
is-a fragment
- IUPAC Name: PHE
Schematic Drawing: 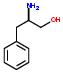
Statement: PHE is-a fragment or  is-a fragment
is-a fragment
- IUPAC Name: 1-benzyl-1-methylhydrazine
Schematic Drawing: 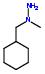
Statement: 1-benzyl-1-methylhydrazine is-a fragment or  is-a fragment
is-a fragment
More examples of IUPAC names of fragments and their schematic drawings can be found in http://xpdb.nist.gov/hivsdb/hiv_ligands.pl and in http://xpdb.nist.gov/hiv2_d/hiv_ligands.pl.
These URLs show the IUPAC names along with the schematic drawings of fragments that have been used to form potent AIDS drugs.
What fragment of given drug has been used in other drugs?
Concepts: Drugs, fragments, chemical linkers. A drug is made up of one or more fragments joined by chemical linkers. In this use case we refer to the drugs with properties to inhibit the biological function of HIV protease – a protein that is crucial for the replication of AIDS virus. All the fragments of drugs used in our use case are generated by breaking at the chemical linkers[6]. Chem-BLAST[6] is used to organize these fragments and to provide answers to the above question. InChI[5] is used to index fragments in the database.
- Example:
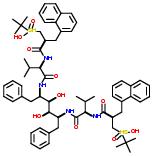
Commercial name of drug: Ritonavir
Schematic Drawing: 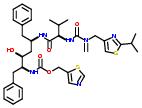
Schematic Drawing of Fragments:
1:2:
3:
4:
5:
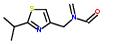
Statements:
1:has-fragment
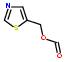
2:has fragment
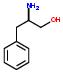
3:has fragment
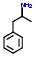
4:has fragment
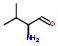
5:has fragment

- Example:
Commercial name of the drug: KNI-272
Schematic Drawing: 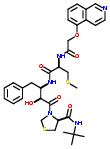
Schematic Drawing of fragments:
Statements:
2:has-fragment
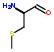
A full list of fragments of AIDS potent drugs may be found in the URLs: http://xpdb.nist.gov/hivsdb/hiv_ligands.pl http://xpdb.nist.gov/hiv2_d/hiv_ligands.pl http://xpdb.nist.gov/hiv2_d/hivsdb_fragment_search.pl http://xpdb.nist.gov/hivsdb/hivsdb_fragment_search.pl
Complex Questions Asked by the Structural Biologist and Modeler
Often many questions are not elmentary. For this reason, we define "conjugated" statements. A conjugated statement is a statement formed by concatenating and/or intersecting more than one statement. For instance, the ‘has common fragment’ statement uses the intersection of two or more ‘has a fragment’ statements.
For example: KNI-272 and Ritonavir has a common fragment shown by the schematic drawing: 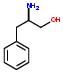
Concepts: Structurally related, structurally identical. Two drugs are considered structurally related when one or more of their fragments are the same. When all the fragments are common between two drugs, they are considered structurally identical or a complete subset.
A ‘Structurally related’ statement is a conjugated statement made by determining the intersection of the ‘has-fragment’ statements of two compounds. If all the fragments from a given drug have an intersection with a fragment of another drug and vice versa, then the two drugs are identical. If only some fragments of a drug has an intersection with the fragments of another drug then the two drugs are similar. The number of such intersections between two drugs may determine the degree of similarity between them. The actual fragments that lead to intersection may shed light on chemical or biological properties such as solubility and drug resistance. Similarity between two compounds may also be defined at different levels of abstraction using class or subgroups discussed later in this document.
Medicinal Chemists View
Questions asked by Medicinal Chemists
Often medicinal Chemists and structural Biologists may have different views of a drug. While structural Biologists may be thinking of the drugs in terms of fragments that provide complementary interaction to the enzyme at specific structural pockets (cavities, refer fig 1 above) medicinal chemists may think of compounds in terms of type of the rings and atoms contained in that drug. The following are some examples of the Medicinal Chemists view.
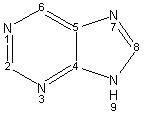
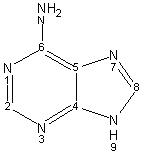
Concepts: Class, subgroup, fragment. Fragments are grouped into their subgroups and subgroups are grouped into class. Class defines the type of atoms a subgroup has and subgroup defines the type of the ring structures and scaffolds in a fragment. Fragments define the building blocks of a compound.
Examples:
Subgroup:
Statement: 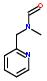 is a fragment
is a fragment
Statement:has subgroup
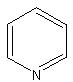
Statement:  has the class nitrogen containing
has the class nitrogen containing
Statement: 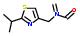 is a fragment
is a fragment
Statement:has subgroup
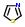
Statement: 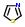 has the class nitrogen containing
has the class nitrogen containing
The statements ‘is a fragment’, ‘has a subgroup’, and ‘has a class’ have relational or taxonomical relationship and hence they are arranged into a data-tree.
Show all the compounds with nitrogen
Example: Displaying the compounds with nitrogen amounts to displaying all the drugs after conjugating ‘has class’, ’has subgroup’, and ‘has fragment’ statements with class = nitrogen containing.
Drugs 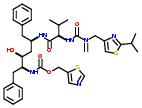 and
and 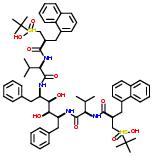 are examples of drugs that belong to the class nitrogen containing.
are examples of drugs that belong to the class nitrogen containing.
To facilitate the conjugation of multiple statements, a chemical data-tree formed by the elements of statements is used. In this data-tree, the subjects and objects of the statements are organized in successive layers as shown below.
Each layer of the data-tree acts a subject for the object defined in the adjacent layers. The predicate that proceeds from top most layer to adjacent layer is ‘has class’, the predicates for the layers that follow the top most layers are ‘has subgroup’,’has fragment’ and ‘is drug’.
Give me all the drugs with a particular subgroup napthyl
Concept: Subgroup, drugs. Drugs are divided into fragments, fragments are divided into subgroups.
IUPAC name: Napthyl
Schematic Drawing: 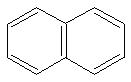
Statement:is subgroup of
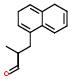
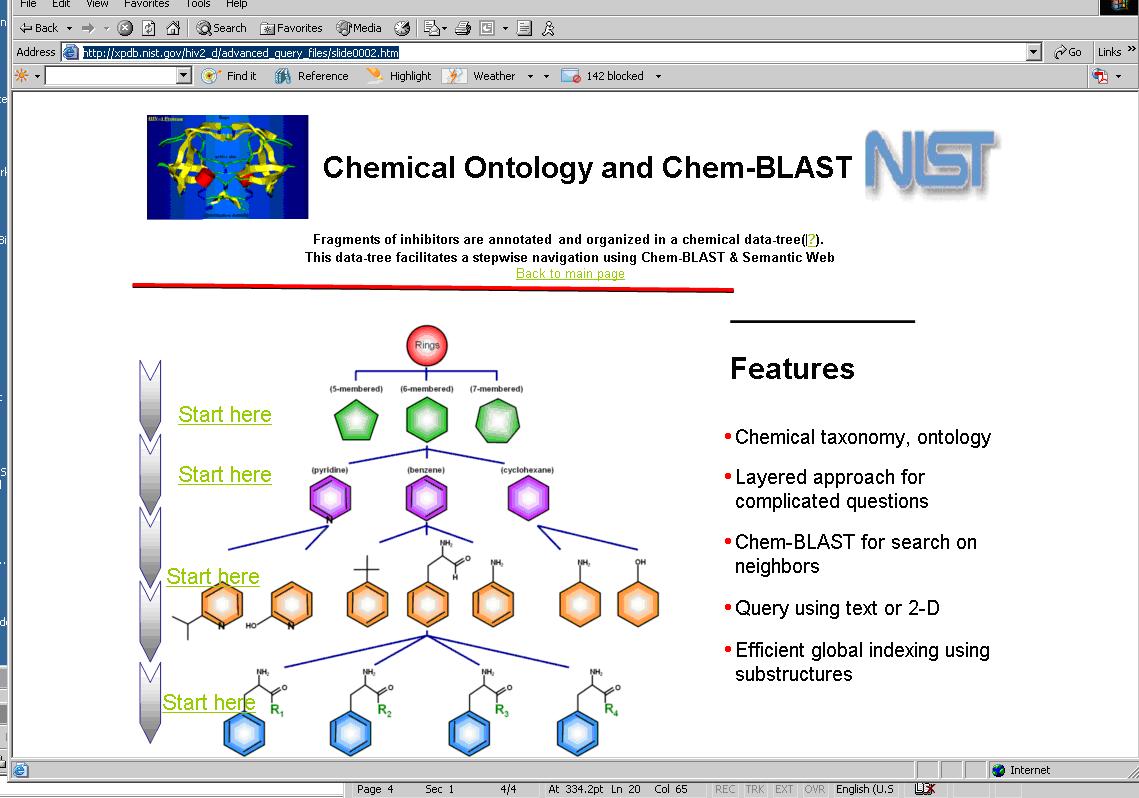
Response to the above question amounts to conjugating the desired ‘is subgroup of’ statement with ‘fragment of’ statement to obtain a list of drugs. Additional examples of this type of use cases may be found in the URLs http://xpdb.nist.gov/hivsdb/advanced_query_files/slide0002.htm and in http://xpdb.nist.gov/hiv2_d/advanced_query_files/slide0002.htm
References
- Drews, J. Drug discovery: a historical perspective. Science 287, 1960 -1964 (2000).
- Kuntz, I. Structure-based strategies for drug design and discovery. Science 257, 1078 - 1082 (1992).
- Rummey, C., Nordhoff, S., Thiemann, M. & Metz, G. In silico fragment-based discovery of DPP-IV S1 pocket binders. Bioorg Med Chem Lett 16, 1405-1409 (2006).
- Blundell, T.L. et al. Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery. Philos Trans R Soc Lond B Biol Sci 361, 413-423 (2006).
- Prasanna, M., Vondrasek, J., Wlodawer, A., Bhat, TN. Application of InChI to curate, index and query 3-D structures. Proteins, Structure, Function, and Bioinformatics 60, 1-4 (2005).
- Prasanna, M.D., Vondrasek, J., Wlodawer, A., Rodriguez, H. & Bhat, T.N. Chemical Compound Navigator: A Web-based Chem-BLAST Search Engine for Browsing Compounds. PROTEINS: Structure, Function, and Bioinformatics 63:907-917 (2006).
A taxonomy based Semantic Web use case for compounds common to HIVSDB and PubChem
Goal
This use case is designed to illustrate the use of a Chemical taxonomy with OWL to annotate, store and query structures that are common to HIV Structural Database (HIVSDB http://xpdb.nist.gov/hiv2_d/hivsdb.html) and PubChem (http://pubchem.ncbi.nlm.nih.gov/).
What is a Chemical Taxonomy?
A chemical taxonomy is the classification of chemical structures according to their relationships using commonly used concepts in the field. The final product of the classification of compounds into a chemical taxonomy is a database and an OWL knowledge base that explicitly relates the compounds using the commonly used concepts. An information resource developer may use the taxonomy to develop Web pages that guide taxonomy searches. A user may use the taxonomy to interactively compose and refine queries to the information resource.
Relationship between OWL and chemical taxonomy
The chemical taxonomy classifies chemicals using certain structural relationships. These relationships may be single layered (i.e., each relationship directly associates commonly used concepts), for example, the groups:
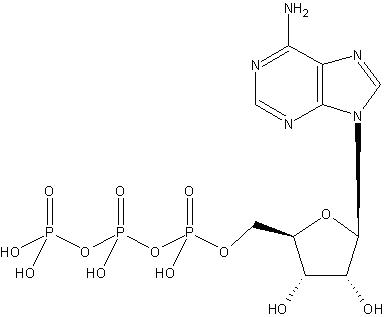
can be associated with a complete compound, such as, ATP (this molecule is source of energy for human body and an average body processes a huge amount of this compound every day):
In this scenario, the groups and the compound become the subject and object of an OWL expression. Alternatively, these relationships may be multi-layered, namely, the commonly used concepts themselves have layered relationships and a concept relates to a compound through one or more other concepts. Some of the concepts do not have direct OWL relationships to the compound. In this scenario, successive related concepts become subject and object of OWL expressions. In the latter case a concept may become a subject of an object, which itself is a subject of another object of an OWL expression.
How one uses a chemical taxonomy for annotation
An annotator first establishes a list of all the commonly used concepts in a given context. Some of these concepts are explained earlier (see Structural Biology and Modeling View and Medicinal Chemists View above). Following this, an annotator organizes each of the concepts such that their inter-relationships, if any, are explicitly defined. For instance, the concepts represented by Pyrimidine and Imidazole may be concepts within the fused ring Purine which is a concept within the compound Adenine. A compound may itself be a concept within another compound (ATP). These individual compounds are analyzed during the annotation step to establish commonly used concepts for the compounds of interest. Different views may be used to define concepts of particular interest to a particular application as described in sections Structural Biology and Modeling View and Medicinal Chemists View.
These concepts are then used to develop the chemical taxonomy. This step involves examination of all the concepts and their relationships and then classifying them into a hierarchy. For instance, the concept represented by Pyrimidine and Imidazole may be defined as instances at the same hierarchical level, since they both define concepts of similar nature. These concepts may be assumed to precede the concept of Purine. The concept of Pyrimidine may be assumed to precede the concept of Purine. The concept of Purine precede that of ATP.
Advantages of chemical taxonomy for annotation
A chemical taxonomy is the organization of the concepts described above into a logical data-tree of several branches and nodes so as to enable browsing and searching. The use of a chemical taxonomy allows annotators to stream-line their thoughts in terms of the commonly used concepts in the field and to express these thoughts in machine logic that may be used during a guided query. This approach reduces ad hoc application of rules during the annotation step, and thus allows the establishment of metrics and standards across the entire information resource system. It also allows expert annotators and the information resource providers to directly impart their knowledge to their customers through the annotation step. In this approach the search engine is ‘data aware’ and it uses individual concepts of the taxonomy to define the exact relationships between different compounds in the context of a user query.
How one uses a taxonomy for database development
A database developer uses the taxonomy to design and populate information about the compounds into data tables. Each layer of the taxonomy may be considered to be a column of the data table with total number of columns equal to the total number of layers of concepts used to define the taxonomy. Alternatively, one may have several data tables with two columns per table; one for the concept and another for the concept that precedes it. The actual choice the structure of the tables may depend on the complexity and dynamic nature of the concepts. Alternatively one may also use a mixture of these two methods to construct the data tables.
Advantage of chemical taxonomy for database development
A chemical taxonomy allows a database developer to accurately define and cross-index concepts within the resource. Using these indices search engines may be developed to facilitate gradual build of a user query starting from commonly used concepts in the field.
How a user queries a chemical taxonomy based Webpage
A search engine displays a graphic of the chemical taxonomy(http://bioinfo.nist.gov/SemanticWeb_hiv2d/chemblast.do and http://xpdb.nist.gov/hiv2_d/advanced_query_files/slide0002.htm) that shows each layer of the taxonomy (concept of a given hierarchy) as an entry point. Users access one of these hyperlinks to begin their query on the concepts represented by that layer. The relationship between these layers can be expressed in OWL.
Advantages of a chemical taxonomy for users
A chemical taxonomy provides a powerful way for an information resource developers to impart their knowledge enabling. An information resource developer organizes his thoughts into individual concepts and establishes relationships among them using OWL. Elements of the OWL representation are used by search engines to guide users in performing their queries as a series of steps - the first step being general and succeeding steps providing an opportunity to refine queries. Using the initial chemical taxonomy graphic, a user may initiate his query starting with rings, e.g., six member ring, then view the resulting choices: Pyrimidine and Imidazole. Then the user may select a Pyrimidine. Subsequently, the user may select Purine and then Adenine. Finally, the user may decide to select ATP. In each step a user gets an opportunity to define his query with greater precision to eliminate unwanted hits. The fact that the user gets to see all the options in each step, also reduces the possibility of missed hits.
How to use the taxonomy to obtain data from PubChem
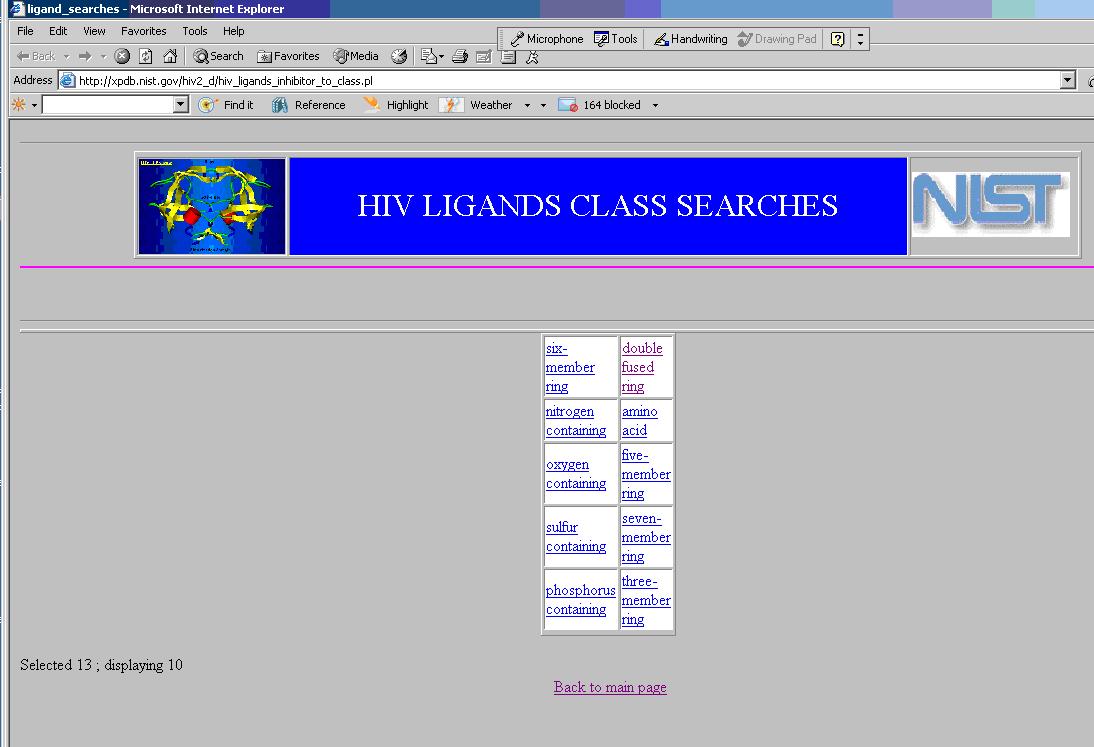
At this time, the chemical taxonomy based search engine exists only in the NIST database. Users perform their initial queries using the chemical taxonomy graphic (at http://bioinfo.nist.gov/SemanticWeb_hiv2d/chemblast.do and http://xpdb.nist.gov/hiv2_d/advanced_query_files/slide0002.htm) in order to access the 2500 compounds that are common to HIVSDB and PubChem.
From this initial page, one may select the top leftmost ‘Start here’ to obtain the Web page shown below to query the class of compounds described in the OWL representation.
Here, the user sees the first layer of the OWL representation. If users now select the second, third, or fourth layers, they will see the corresponding data elements. Each of these layers correspond to particular level of the taxonomy represented by chemical taxonomy. Adjacent layers of the taxonomy are connected by relationships defined in OWL statements.
From the Web page shown above, a user may select a particular type of structure, for instance a double fused ring . Then the following Web page appears that has all the fused rings in the database and denoted by the OWL representation.
Media:HCLS$$ChemicalTaxonomiesUseCase$fig-uc2-7.JPG
From the Web page above, a user may make the next selection, for instance for a quinoline. On making this selection, the Web page will display the fragments that have quinoline as show below.
Media:HCLS$$ChemicalTaxonomiesUseCase$fig-uc2-8.JPG
From here, a user may select one of the fragments, for instance 45222_chain1_5. This will lead to the page shown below . This page has the structures that satisfy all the elements chosen in the previous selections.
Media:HCLS$$ChemicalTaxonomiesUseCase$fig-uc2-9.JPG
Having reached this point, namely the last layer of the chemical taxonomy, a user has several options to look at the data. One of the options in this Web page takes a user to the PubChem Web page for the particular compound as shown below.
Media:HCLS$$ChemicalTaxonomiesUseCase$fig-uc2-10.JPG
As illustrated above, OWL statements are used to form a chemical taxonomy and this taxonomy is used to remotely query the compounds in the PubChem in an intuitive manner to enable gradual build up of query probes. The query probes use OWL statements to connect up chemical structural concepts to guide a user thorough the multiple options available at each stage of the query.