Abstract
One of the challenges facing Semantic Web for Health Care and
Life Sciences is that of converting relational databases into
Semantic Web format. The issues and the steps involved in such a
conversion have not been well documented. To this end, we have
created this document to describe the process of converting
SenseLab databases into OWL. SenseLab is a collection of relational
(Oracle) databases for neuroscientific research. The conversion of
these databases into RDF/OWL format is an important step towards
realizing the benefits of Semantic Web in integrative neuroscience
research. This document describes how we represented some of the
SenseLab databases in Resource Description Framework (RDF) and Web
Ontology Language (OWL), and discusses the advantages and
disadvantages of these representations. Our OWL representation is
based on the reuse of existing standard OWL ontologies developed in
the biomedical ontology communities. The purpose of this document
is to share our implementation experience with the community.
Appendices
Conversion process
Original data sources
The SenseLab databases can be accessed through a web interface
at the SenseLab website [ SENSELAB-WEB ]. SenseLab is divided into a
number of specialised databases, of which we have converted three
to Semantic Web formats. These databases are NeuronDB, BrainPharm
and ModelDB. All databases are based on compartmental models of
neurons. NeuronDB contains descriptions of anatomic locations, cell
architecture and physiologic parameters of neuronal cells. The
pilot BrainPharm database is intended to support research on drugs
for the treatment of neurological disorders. It enhances the
descriptions in a portion of NeuronDB with descriptions of the
actions of pathological and pharmacological agents. ModelDB is a
large repository of computational neuroscience models and
simulations. The mathematical models in ModelDB are annotated with
references to NeuronDB. Taken together, these databases allow the
researcher to query information and to run simulations pertaining
to the function of neurons in healthy and disease states. All
databases contain extensive literature references and excerpts from
texts that have been used to curate the database entries.
The databases are based on the "entity-attribute-value with
classes and relationships" (EAV/CR) schema [ EAV-CR ]. The data can also be downloaded from
the SenseLab Semantic Web developments portal [ SENSELAB-SW ] as a database dump in
Microsoft Acces MDB format and as text.
Initial RDF and OWL conversions
Motivation
Our motivation was to make the SenseLab databases available in
RDF [ RDF ] (without OWL) and in OWL DL [
OWL Overview ]. The two versions
were developed in parallel in order to compare the difference
between the conversion processes and the outcomes. We wanted to
explore the issues in mapping relational databases to RDF/OWL
structure. In addition, we wanted to explore the possibility of
automatic translation from EAV/CR to RDF.
Process
We developed a converter application in Java that queried the
SenseLab database and wrote RDF/XML files. The conversion was fully
automatic for the RDF version, but required some manual editing for
the OWL version.
Outcome
These conversions were too tied to the original database
structure, which resulted in inconsistent OWL ontologies. Some
shortcomings of the first conversion to OWL were:
- 'Part of' relations were incorrectly represented as subclass
relations. This seems to be one of the most common mistakes in
ontology development in general.
- Class disjoints ¹ were missing which made it hard to find
inconsistencies and data entry errors.
- After disjoints were introduced, we found some previously
unidentified inconsistencies with the help of OWL reasoners: some
classes (e.g. 'GABA') were subclasses of both 'neurotransmitter'
and 'receptor', which was wrong. This was an artifact caused by the
automated conversion -- both GABA transmitters and GABA receptors
were simply labeled with 'GABA' in the source database. The
conversion algorithm generated URIs based on these labels, so they
were represented identical URIs (
http://neuroweb.med.yale.edu/senselab/neuron_ontology.owl#
GABA ). This grave mistake would not have
been noticed without the use of OWL reasoning.
- Some of the labels of entities were very terse and not
understandable outside the user interface of the original database.
For example, "Ded" was the label of the "distal part of the
dendrite".
¹ Disjoint classes are
used in OWL to assert that they have no members in common.
Inferences from this can be used to flag an inconsistent
models.
Revised OWL conversions
The revised OWL conversion was based on the first OWL
conversions. The design of the SenseLab ontologies follows the
"ontological realism" approach [ SMITH-2004 ]. This means that the ontologies
are focused on direct representations of physical objects and
processes (e.g., neuronal cells, ionic currents), and not on their
abstractions (e.g., concepts or database entries).
Motivation
Manually correcting the logical inconsistencies in the first
version of the OWL ontology; making use of foundational ontologies
(BFO, Relation Ontology) where possible; mapping the ontology to
other neuroscience ontologies.
Process
An ontology containing basic class hierarchies and relations was
manually created, based on the structure of existing SenseLab
databases. This basic ontology could not be created from the
database structure in an automated process because this would not
have resulted in a logically consistent ontology. This ontology was
edited by a domain expert, based on inspection and manual editing
with Protege 3.2 [ PROTEGE ] and
Topbraid Composer [ TOPBRAID ]. The
ontologies were built upon established foundational ontologies in
order to maximize the interoperability with other existing and
forthcoming biomedical Semantic Web resources. These ontologies
were:
- the Relation Ontology [ RO ] from the
Open Biomedical Ontologies repository [ OBO
], which defines basic relations such as 'part of', 'participant
of' or 'contained in'.
- the Basic Formal Ontology [ BFO ], which
defines basic classes such as 'process', 'object', 'quality' or
'function'.
Based on this manually created basic ontology, the data from the
SenseLab databases were then automatically converted to OWL using
programs written in Java and Python. The automated export scripts
extended the manually created basic ontology through the creation
of subclasses, OWL property restrictions and individuals. The
resulting ontologies show no clearly distinguishable divide between
a 'schema' and 'data'.
The OWL export of NeuronDB was based on a transformation from
the EAV/CR model of the SenseLab database to RDF/XML by a Java
program. The export from ModelDB and BrainPharm was based on a
simple flat text file export of the databases. The text file
exports were converted to RDF/XML files with a Python script.
For mappings to external bioinformatics databases that do not
yet offer stable URIs for reference on the Semantic Web, we used
the URI scheme for database record identifiers established by
Science Commons [ SC-URI ]. URIs for
database records could simply be generated by concatenating the
record identifier to a predefined namespace. For example, the
Entrez Gene record with ID '3579' was identified by the URI
http://purl.org/commons/record/ncbi_gene/
3579 , the Uniprot record 'P46663' was
identified by http://purl.org/commons/record/uniprotkb/
P46663 and the Pubmed record with ID
'11160518' was identified by http://purl.org/commons/record/pmid/
11160518 . The database entries were
connected to the ontological representations of real-word entities
through relations such as
has_nucleotide_sequence_described_by . For example,
the gene of the Dopamine Receptor D1 (DRD1) is defined through a
reference to NCBI record 1812, which contains a description of the
sequence of this specific gene:
<http://purl.org/ycmi/senselab/neuron_ontology.owl#DRD1_Gene>
owl:equivalentClass _:property_restriction1 .
_:property_restriction1 owl:onProperty
senselab:has_nucleotide_sequence_described_by .
_:property_restriction1 owl:hasValue
<http://purl.org/commons/record/ncbi_gene/1812> .
Mappings were made to the following ontologies:
- the BAMS ontology which was derived from the Brain Architecture
Management System [ BAMS ]
- the Subcellular Anatomy Ontology (SAO) created by the Cell
Centered Database project. [ SAO ]
- the BirnLex ontology developed by members of the Biomedical
Informatics Research Network [ BIRNLEX
]
- the Common Anatomy Reference Ontology (CARO)
[ CARO ]
- the Gene Ontology [ GO ]
- the Ontology of Biomedical Investigation (OBI) [ OBI ]
The mappings were made with the following cross-ontology
relations: owl:equivalentClass
, rdfs:subClassOf
and the "has part"
relation from the OBO relation ontology .
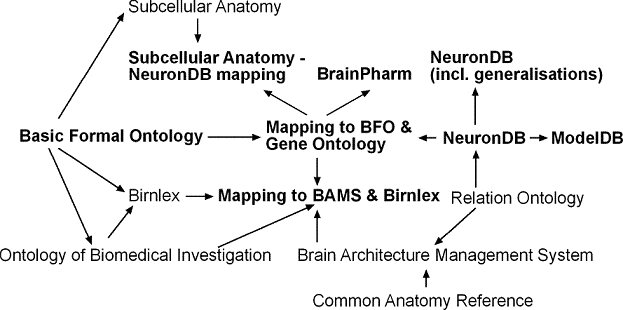
Figure 1: Import hierarchy of OWL ontologies. Ontologies printed
in bold have been created by the SenseLab team, other ontologies
have been created by other groups. The arrows point from the
imported ontology to the importing ontology, e.g., the NeuronDB
Ontology imports the Relation Ontology. Import statements are
transitive, e.g., the ModelDB Ontology imports both the NeuronDB
ontology and the Relation ontology.
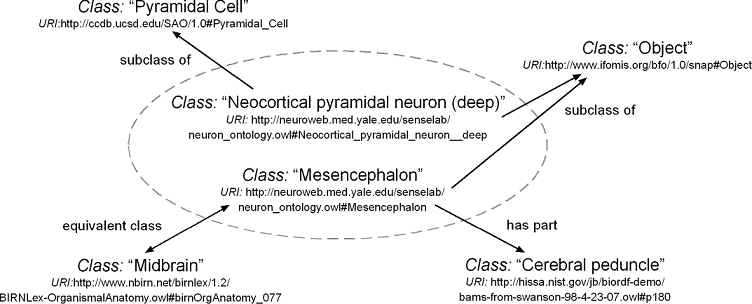
Figure 2: Examples of relations ('mappings') spanning between
classes from the NeuronDB ontology (in the middle) and classes from
external ontologies.
Terse rdfs:labels were replaced by more descriptive ones that
could be better understood without knowledge about context. For
example, the rdfs:label "Ded" was changed to "Distal part of
equivalent dendrite (Ded)". Note that, in this case, the original
label was also preserved (in brackets), because it might still be
useful for people that do know about the context.
The ontology development was moved to a Subversion (SVN) system
on a central webserver. During most of the development, the
ontologies were simply developed on the client side and were
periodically uploaded via FTP. Of course this led to problems when
more than one person was working on the ontologies at a time, and
it was also impossible for users of the ontology to access previous
versions of the ontology, since only the most recent version was
available on the website.
The namespaces / ontology locations were changed to PURL-based
URIs. For example, the URI http://
neuroweb.med.yale.edu
/senselab/neuron_ontology.owl#Dopamine was changed to
http://
purl.org/ycmi
/senselab/neuron_ontology.owl#Dopamine . PURL-based URIs
are easier to maintain when server configurations change or (in the
worst case) the original server is unavailable and the ontologies
need to be served from a different location. The increased
stability of PURLs encourages the re-use of entities in ontologies
developed by other groups -- which is a key factor in the creation
of a coherent Semantic Web.
A SPARQL endpoint for the SenseLab ontologies was set up using
the open source version of the Openlink Virtuoso server [ VIRTUOSO ]. A SPARQL endpoint is a service that
allows clients to query a RDF store with the SPARQL query language
through simple HTTP GET requests. The ontologies were loaded into
the triplestore of the server to make them accessible to SPARQL
queries. Each ontology file was put into a separate labeled graph,
the label of each graph was identical to the URL of the ontology
file. For example, the ontology located at http://purl.org/ycmi/senselab/neuron_ontology.owl
was loaded into a graph labeled
http://purl.org/ycmi/senselab/neuron_ontology.owl .
Loading each ontology into a separate graph makes it possible to
restrict SPARQL queries to certain graphs and hence, certain
ontologies. This has the advantage that queries can be more
selective and can be executed with better performance.
Outcome
The final products of the project are accessible at http://neuroweb.med.yale.edu/senselab/
. A Subversion (SVN) repository can be accessed through a web
interface at http://neuroweb.med.yale.edu/svn/trunk/ontology/senselab/
. The SPARQL endpoint can be accessed at http://hcls.deri.ie/sparql . The
SenseLab OWL ontologies are mentioned as a primary example for the
application of OBO ontologies in the article The OBO Foundry:
coordinated evolution of ontologies to support biomedical data
integration [ OBO-ARTICLE
].
Advantages
The use of OWL significantly eased the integration of SenseLab
data with ontologies developed by other projects. OWL-based data
integration does not require the development and maintenance of
central mediators, reducing development and maintenance costs. The
ontology integration can be accomplished by creating meaningful
relations between entities in distributed ontologies.
Ontologies can be modularized; dependencies between ontologies
can be made explicit through 'owl:imports' statements. This makes
distributed development of ontology modules feasible and encourages
the re-use of selected ontology modules by other groups.
Good OWL ontologies are self-descriptive because every entity
can be annotated with text.
Reasoners can be used to identify errors and real (i.e.,
conscious) contradictions in submitted data sets. You might find
more errors and contradictions than you expected.
Ontologies can be used to directly represent biological reality
without introducing unnecessary abstractions such as database
tables, data dictionaries, and documents.
Disadvantages
The open-source ontology editors used for this project were
relatively unreliable. A lot of time was spent with steering around
software bugs. Future versions of freely available editors (e.g.
Protege 4) or currently available commercial ontology editors (e.g.
Topbraid Composer) might be preferable.
Descriptions of OWL classes and their relations (i.e., OWL
property restrictions) result in very complex and unintuitive RDF
graphs. This makes it hard to generate them automatically, or to
write SPARQL queries that query such ontologies.
Current reasoners can still have performance problems when
checking / classifying complex OWL ontologies.
The RDF/XML serialisation of RDF is not very easy to work with.
It is often a source of errors.
Future directions and plans
Future OWL conversions are planned to be based on the
intermediate, syntactic RDF conversion. The SenseLab ontologies
will be further integrated with other neuroscientific and
biomedical ontologies. User friendly applications will be developed
to query a multitude of interrelated ontologies in a scientifically
meaningful way.
Suggestions based on our experiences
Try to create consistent OWL DL ontologies. Pure RDF(S) without
OWL constructs is not much simpler than OWL DL and often leads to
the creation of too many properties because pure RDF(S) does not
support property restrictions.
Try to re-use entities and properties from existing ontologies
where possible.
If you do not want to import another ontology in its entirety
(e.g. because it would be too large, too buggy or would introduce
unnecessary constructs), you can still 'copy & paste' portions
of the ontology into your own.
Try to base your ontology on a foundational ontology like BFO,
OBO Relation Ontology or DOLCE [ DOL ].
Where possible use the rdfs:label property to give clear,
understandable to each entity and property in the ontology. Try to
formulate labels in a way that makes them understandable without
too much additional context (e.g. a certain user interface).
Where possible, give concise rdfs:comments.
Make a habit out of running your ontology through the RDF
validator [ RDF-VALID ] periodically,
especially when you create RDF/XML with scripts that you wrote
yourself. Keep in mind that the RDF validator does not throw an
error message when URIs contain blanks. Blanks in URIs are
problematic for many Semantic Web applications, so try to make sure
that your URIs do not contain blanks.
Check the consistency of your OWL ontology periodically. We used
the Pellet reasoner [ PELLET ], which
seems to be the best choice at the moment.
Use purl.org URIs for your ontologies. You can easily register a
sub-domain at purl.org free of charge.
If you write a program that generates RDF/OWL, do
NOT try to write RDF/XML code directly. RDF/XML is
relatively complicated and messy, and it is very easy to produce
syntactic or even semantic errors because of that. So if you write
a program that generates RDF, use a RDF or OWL API for writing
triples. If that is not possible, generate your RDF in the much
simpler TURTLE syntax instead of RDF/XML. The TURTLE syntax is a
subset of the N3 syntax [ N3 ]. You can save
the resulting RDF in TURTLE format to a text file. If you need
RDF/XML for another application, you can convert the TURTLE to
RDF/XML in a second step.
References
- [EAV-CR]
- L. Marenco, N. Tosches, C. Crasto, G. Shepherd, P.L. Millera
and P.M. Nadkarni, Achieving evolvable Web-database bioscience
applications using the EAV/CR framework: recent advances, J Am Med
Inform Assoc. (2003) 10(5):444-53
-
[SENSELAB-WEB]
- SenseLab database ,
http://senselab.med.yale.edu/
-
[SENSELAB-SW]
- SenseLab
Semantic Web Development ,
http://neuroweb.med.yale.edu/senselab/
- [PROTEGE]
- The Protege Ontology
Editor and Knowledge Acquisition System ,
http://protege.stanford.edu/
- [TOPBRAID]
- TopBraid Composer ,
http://www.topbraidcomposer.org/
- [RO]
- Relation
Ontology , http://www.obofoundry.org/ro/
- [OBO]
- The Open Biomedical
Ontologies , http://obofoundry.org/
- [BFO]
- Basic
Formal Ontology (BFO) ,
http://www.ifomis.uni-saarland.de/bfo/
- [SC-URI]
- Explanation
of HCLS and Science Commons URIs ,
http://sw.neurocommons.org/2007/uri-explanation.html
- [BAMS]
- The Brain
Architecture Management System ,
http://brancusi.usc.edu/bkms/
- [SAO]
- CCDB
Subcellular Anatomy Ontology ,
http://ccdb.ucsd.edu/CCDBWebSite/sao.html
- [CARO]
- Common
Anatomy Reference Ontology ,
http://www.obofoundry.org/cgi-bin/detail.cgi?id=caro
- [BIRNLEX]
- BIRNLex Ontology Documentation ,
http://fireball.drexelmed.edu/birnlex/OWLdocs/
- [GO]
- Gene Ontology ,
http://geneontology.org/
- [OBI]
- Ontology of Biomedical
Investigation , http://obi.sourceforge.net/
- [VIRTUOSO]
- OpenLink Universal
Integration Middleware - Virtuoso Product Family ,
http://virtuoso.openlinksw.com/
-
[OBO-ARTICLE]
- The OBO Foundry: coordinated evolution of ontologies to
support biomedical data integration , Barry Smith, Michael
Ashburner, Cornelius Rosse, Jonathan Bard, William Bug, Werner
Ceusters et al. , Nature Biotechnology 25, 1251 - 1255,
2007, http://dx.doi.org/10.1038/nbt1346
- [DOL]
- DOLCE
Ontology , http://www.loa-cnr.it/DOLCE.html
-
[RDF-VALID]
- RDF
Validator , http://www.w3.org/RDF/Validator/
- [PELLET]
- The PELLET Open Source
OWL DL Reasoner , http://pellet.owldl.org/
-
[SMITH-2004]
- Beyond Concepts: Ontology as Reality Representation ,
Barry Smith, iin A. Varzi, L. Vieu, eds., Proceedings of FOIS (IOS
Press, Amsterdam, 2004) 319-330. http://ontology.buffalo.edu/bfo/BeyondConcepts.pdf
- [KB]
HCLS Knowledgebase A Prototype
Knowledge Base for the Life Sciences , http://www.w3.org/TR/2008/WD-hcls-kb-20080404/
http://www.w3.org/TR/2008/NOTE-hcls-kb-20080604/- [N3]
- Primer:
Getting into RDF and Semantic Web using N3 ,
http://www.w3.org/2000/10/swap/Primer
- [OWL
Overview]
- OWL Web
Ontology Language Overview , Deborah L. McGuinness and
Frank van Harmelen, Editors, W3C Recommendation, 10 February 2004,
http://www.w3.org/TR/2004/REC-owl-features-20040210/ .
Latest version available at http://www.w3.org/TR/owl-features/
[RDF] Resource Description Framework (RDF) Model and Syntax
Specification , Ora Lassila, Ralph R. Swick, Editors. World Wide
Web Consortium Recommendation, 1999,
http://www.w3.org/TR/1999/REC-rdf-syntax-19990222/. Latest version
available at http://www.w3.org/TR/R http:/

