Abstract
Developing cures for highly complex diseases, such as neurodegenerative disorders, requires extensive interdisciplinary collaboration and exchange of biomedical information in context. Our ability to exchange such information across sub-specialties today is limited by the current scientific knowledge ecosystem’s inability to properly contextualize and integrate data and discourse in machine-interpretable form. This inherently limits the productivity of research and the progress toward cures for devastating diseases such as Alzheimer’s and Parkinson’s. The SWAN (Semantic Web Applications in Neuromedicine) ontology is an ontology for modeling scientific discourse and has been developed in the context of building a series of applications for biomedical researchers, as well as extensive discussions and collaborations with the larger bio-ontologies community. This document describes the SWAN ontology of scientific discourse.
Status of This Document
This section describes the status of this document at the time of its publication. Other documents may supersede this document. A list of current W3C publications and the latest revision of this technical report can be found in the W3C technical reports index at http://www.w3.org/TR/.
This W3C Interest Group Note describes how one can use the Semantic Web to express and integrate scientific data. These techniques can be used for modeling any data, and the benefits of integration and model consistency apply to other diverse, distributed data domains. It is hoped that this document will inspire further contributions to the ongoing work at Neurocommons and the Health Care and Life Sciences Interest Group, as well as inspire those in other domains to exploit the Semantic Web.
This document describes the SWAN ontology for expressing scientific discourse. The companion document SIOC, SIOC Types and Health Care and Life Sciences describes the SIOC ontology for semantically-interlinked online communities, and SWAN/SIOC: Alignment Between the SWAN and SIOC Ontologies describes the the use of SWAN and SIOC together to model discourse within scientific communities.
The document was produced by the Semantic Web in Health Care and Life Sciences Interest Group (HCLS), part of the W3C Semantic Web Activity (see charter). Comments may be sent to the publicly archived public-semweb-lifesci@w3.org mailing list.
Publication as an Interest Group Note does not imply endorsement by the W3C Membership. This is a draft document and may be updated, replaced or obsoleted by other documents at any time. It is inappropriate to cite this document as other than work in progress.
The disclosure obligations of the Participants of this group are described in the charter.
Appendices
1 Introduction
In many formal models of knowledge acquisition in science, research proceeds in a cycle – from hypothesis development; through experiment and data collection; to interpretation and drawing of conclusions; to communication of results to other scientists; to assimilating, criticizing and synthesizing the communications of colleagues. These practice-theory-practice cycles are socially interconnected in an extremely rich and complex way in what has been termed the “knowledge ecosystem” of science. More and more this knowledge ecosystem is mediated by the technology of the Web. Philosophers of science have defined knowledge as “warranted true belief” [ENCY-PHIL]. The classical knowledge management definition of knowledge is a bit more limited: “information in context” [WORKING-KNOWLEDGE]. This latter definition is insufficiently specific about evidence and the material basis of knowledge. Scientific knowledge strives to approximate objective truth, about a world that exists independently of our beliefs about it. Therefore scientific knowledge by its nature, requires experimental validation - evidence - as a warrant for belief. For scientific knowledge management systems, the context of information is its warrant for belief, while experiment in relation to theory and hypothesis supplies the criterion of truth. Discourse and social practice (of which it is a part) weave this whole together. The aim of the SWAN ontology [SWAN-ONT-JBI] is to enable a social-technical ecosystem in which semantic context of scientific discourse can be created, stored, accessed, integrated and exchanged along with unstructured or semi-structured digital scientific information. The SWAN ontology is freely accessible on the web [SWAN-ONT-WEB]. and provides a formal basis in OWL [OWL] for organizing a very rich context for scientific information and discussion. The SWAN ontology, which was originally monolithic, has been modularized to foster re-usability and integration with other existing ontologies. Thus, when we refer to the SWAN ontology, we actually refer to a collection of ontologies (or modules).
1.2 Document Scope and Target Audience
This document attempts to succinctly describe how the SWAN ontology (version 1.2) was constructed so that interested parties can use all or parts of it to create their own knowledge base. Particular attention will be dedicated to the representation of the scientific discourse. Those interested in more detailed information about the other ontology modules see SWAN Ontology Ecosystem.
1.3 Document Conventions
The following namespace prefix bindings are assumed unless otherwise stated:
| Prefix | URI | Description |
|---|
rdf: |
http://www.w3.org/1999/02/22-rdf-syntax-ns# | The RDF Vocabulary |
rdfs: | http://www.w3.org/2000/01/rdf-schema# | The RDF Schema vocabulary |
xsd: | http://www.w3.org/2001/XMLSchema# | XML Schema |
foaf: | http://xmlns.com/foaf/0.1/ |
Friend Of A Friend |
skos: | http://www.w3.org/2008/05/skos# |
Simple Knowledge Organization System |
dc: | http://purl.org/dc/elements/1.1/ |
Dublin Core Metadata Element Set, Version 1.1 |
dcterms: | http://purl.org/dc/terms/ |
Dublin Core Metadata Terms |
The following SWAN modules namespace prefix bindings are assumed unless otherwise stated:
swande: |
http://purl.org/swan/1.2/discourse-elements/ |
Discourse Elements |
swandr: |
http://purl.org/swan/1.2/discourse-relationships/ |
Discourse Relationships |
swanpav: |
http://purl.org/swan/1.2/pav/ |
Provenance, Authoring and Versioning |
swanqs: |
http://purl.org/swan/1.2/qualifiers/ |
Qualifiers |
swanco: |
http://purl.org/swan/1.2/swan-commons/ |
Commons |
swanci: |
http://purl.org/swan/1.2/citations/ |
Bibliographic Citations |
swanag: |
http://purl.org/swan/1.2/agents/ |
Agents (extension of FOAF) |
swanq: |
http://purl.org/swan/1.2/qualifiers/ |
Qualifiers |
2 Use Case
The SWAN ontology has been initially developed for covering the requirements of the SWAN Project [SWAN-PROJECT]. The SWAN project aims to develop a practical, common, semantically-structured framework for scientific discourse in bio-medicine in general, and neuro-medicine in particular. The ontology of discourse at the core of SWAN is an ontology about what is said, rather than about agreed- upon objective facts.
Initially applied to significant problems in Alzheimer Disease (AD) research, the SWAN project is the result of collaboration between the Alzheimer Research Forum (Alzforum) and informaticians at Massachusetts General Hospital and Harvard University. The first Implementation has been made available as the SWAN Alzheimer Knowledge Base [SWAN-ALZHEIMER].
A process of integration of the SWAN ontology with the Science Collaboration Framework [SCF] is currently in progress. The Science Collaboration Framework is a project of theMassachusetts General Hospital and the Initiative in Innovative Computing at Harvard University in collaboration with the Harvard Stem Cell Institute. It is designed to support interdisciplinary scientists in publishing, annotating, sharing and discussing content such as articles, perspectives, interviews and news items, as well as assert personal biographies and research interests. These web materials can then be linked to external, heterogeneous knowledge repositories of life science resources such as genes, antibodies, cell-lines or model organisms.
3 Design Decisions
3.1 Modularization
Realizing the full potential of the semantic web requires the large-scale adoption and use of ontology-based approaches for sharing of information and resources. Ontology reuse represents a crucial factor in the pursuit of this goal. Moreover, constructing large ontologies typically requires collaboration among multiple individuals or groups with expertise in specific areas, with each participant contributing only a part of the ontology. Therefore, instead of a single, centralized ontology, in most domains, there are multiple distributed ontologies covering parts of the domain.
The SWAN ontology ecosystem has been created as a set of ontologies or modules. Thus, when we refer to the SWAN ontology, we actually refer to a collection of ontologies or modules. Each module is covering one single topic and is developed to have the highest cohesion and the lowest coupling possible. This modularized approach has been adopted for different reasons:
- small and cohesive ontologies are easier to create, verify, maintain and understand. An iterative development method can be applied and those interested in one single aspect of the ontology can focus on a specific module without having to understand the architecture and details of the entire ecosystem
- modularization allows a partial and independent reuse of single components of the ontology ecosystem. The SWAN ontology has been initially developed for representing scientific discourse in neuromedicine. However, the current architecture allows those interested to adopt a significant part of the SWAN ontology for representing discourse related to any domain in science. And this would still assure an important level of integration with other SWAN ontology-based applications
- modularization allows ontology swapping when needed. Given the high cohesion and the low coupling it is easier to substitute a module with a third party domain ontology covering the same topic.
3.2 Fostering Reuse
Even if, currently, most of the integrated domain ontologies have been developed in the context of the SWAN project to fulfil specific requirements, one of the main objectives of the SWAN project is third parties domain ontologies reuse. In fact, it is not realistic to think of the SWAN project as the definitive provider for all the domain ontologies related to biomedicine and science in general.
The core of the SWAN ontology is represented by the scientific discourse module. This represents the basic infrastructure that can be enriched through integration of domain ontologies. An implementation of the domain ontologies required by the SWAN Alzheimer Knowledge Base is provided [SWAN-ONT-WEB]. In parallel, the community gravitating around the SWAN ecosystem is involved in a constant process of monitoring new and growing domain ontologies developed by other communities. If such ontologies are proven to meet the application requirements and demonstrate a larger adoption than the SWAN modules, they can be integrated into the SWAN ontology ecosystem replacing the existing implementations.
3.3 OWL-DL
The SWAN ontology is written in OWL. The sublanguage OWL-DL has been selected as it offers great expressivity without losing computational completeness (all entailments are guaranteed to be computed) and decidability (all computations will finish in finite time) of reasoning systems. Reasoners are helpful to identify errors and inconsistencies in the ontology and errors and contradictions in the data sets. Moreover, in OWL, ontologies can be modularized and dependencies between ontologies can be made explicit through 'owl:imports' statements. And this is particularly helpful to implement the approach presented in the previous paragraph [3.1].
As not all the ontologies integrated in the SWAN ontology ecosystem are natively expressed in OWL-DL, we defined new versions of them in OWL-DL. This is the case for the Friend of A Friend [FOAF] and Simple Knowledge Organization System [SKOS] vocabularies. The first is originally defined in RDFS [RDFS] and the latter in OWL Full.
4 Architecture
The SWAN ontology is organized in three types of modules:
- basic: basic modules represent the ontology building blocks (see figure 1). They cover topics that are general enough to be included in every ontology distribution. The current basic modules are:
- collections: it defines unordered collections (Set and Bag) and ordered collections (or List). This module has been inspired by the work "Putting OWL in Order: Patterns for Sequences in OWL" by Drummond et al. [OWL-ED 2006]. Ordered collections are necessary for representing, for instance, list of authors and the right order in which discourse elements are organized in a scientific discourse.
- provenance, authoring and versioning (PAV) : it provides all the relationships that we need for defining provenance, authoring and versioning which are not already part of Dublin Core [DC] and Dublin Core Terms [DC-TERMS] vocabularies.
- discourse relationships: these are a set of relationships that can be used to build discourse and in particular scientific discourse. The discourse relationships have been defined without constraining domains and ranges. This allows their reuse in contexts different than the SWAN applications.
- FOAF (in OWL-DL): it describes persons, their activities and their relations to other people and objects such as projects, organizations, groups and so on.
- agents: it is extending the FOAF ontology to cover our requirements (it is dependent from the collections and FOAF modules).
- SKOS (in OWL-DL): it is a family of formal languages designed for representation of thesauri, classification schemes, taxonomies, subject-heading systems, or any other type of structured controlled vocabulary.
- qualifiers: it introduces a mechanism for performing annotation of entities through controlled vocabularies and it is based on SKOS.
- scientific discourse: it introduces classes and relationships for representing the scientific discourse.
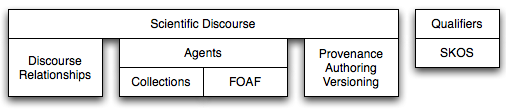
Figure 1: The SWAN ontology basic modules and their dependencies
- extension: extensions modules are covering topics (a) that can be related only to some fields of science (b) for which there could be more than one implementation (c) for which a temporary solution has been provided and it is possible to forecast its substitution. The current extensions modules are:
- life science entities: it is currently dealing with genes, proteins and organisms and relationships in between them. It is covering the SWAN project requirements and it is not of interest for communities that are not related to biomedicine/biology. In the future it will probably be transformed into a connector for organizing different external domain ontologies that will evolve independently from the SWAN ontology
- citations: it covers the requirements of the SWAN applications but could be swapped with another ontology for representing bibliographic records such as the Bibliographic Ontology [BIBO].
- qualifiers extension modules: they are a collection of controlled vocabularies that are used to perform annotation.
- distribution: distributions are modules that are including all the basic modules and extensions necessary for serving a specific domain (i.e. the SWAN ontology for the Alzheimer knowledge base or the generic distribution that is not binded to any specific scientific domain). The current available distributions are:
- swan-scientific-discourse: it includes all the modules we believe can be reused in any scientific context (see figure 2)
- swan-alzheimer: it includes all the modules that are used by the SWAN Alzheimer knowledge base(see figure 3)
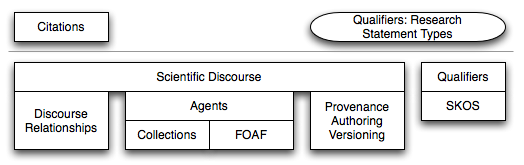
Figure 2: Modules of the SWAN Scientific Discourse distribution.
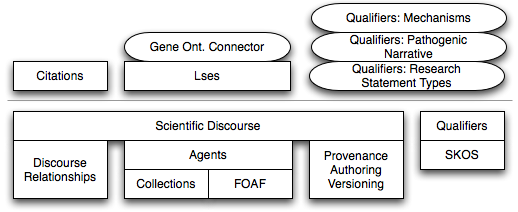
Figure 3: Modules of the SWAN Alzheimer knowledge base distribution.
5 Examples of Discourse Elements
5.1 Claim
The following claim has been extracted from the SWAN Alzheimer Knowledge Base.The SWAN Alzheimer knowledge base is currently using LSIDs as unique identifiers but for demonstrations purposes, and given the fact that also the SWAN project is soon going to migrate to purl-based identifiers, we will use more readable unique identifiers.
Example of SWAN claim instance in Turtle syntax:
@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> .
@prefix swande: <http://purl.org/swan/1.2/discourse-elements/> .
@prefix swanco: <http://purl.org/swan/1.2/swan-commons/> .
@prefix swanqs: <http://purl.org/swan/1.2/qualifiers/> .
@prefix swandr: <http://purl.org/swan/1.2/discourse-relationships/> .
@prefix swanpav: <http://purl.org/swan/1.2/pav/> .
<http://hypothesis.alzforum.org/researchstatement/100> a swande:ResearchStatement ;
swande:title "Aside from its well-established role in promoting the stabilization
of microtubules (MTs), tau may have additional functions as a result of its interactions with
other structures and enzymes"@en;
swande:description "Poorly defined interactions and functions of tau contribute
to the difficulty of understanding how pathologically altered tau mediates neurodegeneration.
For example, tau interacts with the plasma membrane, the actin cytoskeleton and with src
tyrosine kinases such as FYN."@en;
swanco:citesAsSupportiveEvidence <http://hypothesis.alzforum.org/citation/321>,
<http://hypothesis.alzforum.org/citation/322>,
<http://hypothesis.alzforum.org/citation/323>,
<http://hypothesis.alzforum.org/citation/324>,
<http://hypothesis.alzforum.org/citation/325>;
swanco:researchStatementQualifiedAs
<http://swan.mindinformatics.org/ontologies/1.2/rsqualifiers/claim>;
swanqs:qualifiedBy
<http://swan.mindinformatics.org/ontologies/1.2/pathogenic-narrative/initial_condition>;
swandr:refersTo
<http://hypothesis.alzforum.org/protein/201>;
swanpav:curatedBy <http://hypothesis.alzforum.org/people/gwen_wong>;
swanpav:createdBy <http://hypothesis.alzforum.org/people/elizabeth_wu>;
swanpav:createdOn "April 1, 2009".
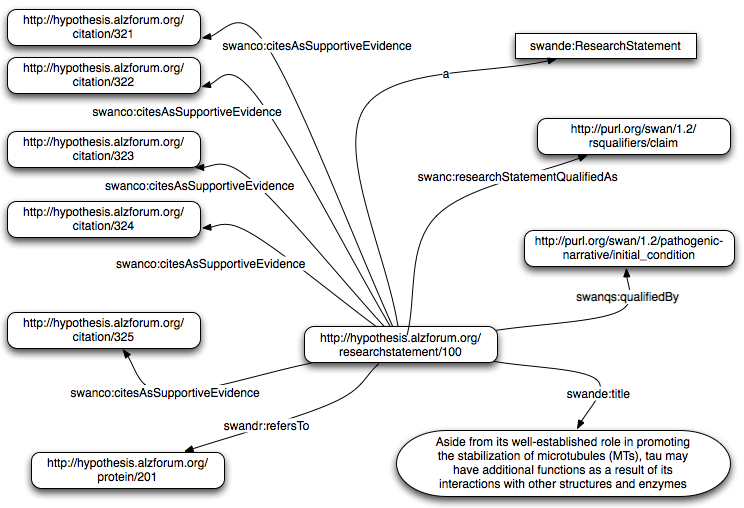
Figure 4: Example of claim.
5.2 Hypothesis
The following hypothesis has been extracted from the SWAN Alzheimer Knowledge Base and refers (or contains, according to the SWAN discourse relationships module) to the claim of the previous example as part of its discourse. The number of referred statements has been reduced to increase readability.
Example of SWAN hypothesis in Turtle syntax:
@prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> .
@prefix swande: <http://purl.org/swan/1.2/discourse-elements/> .
@prefix swanco: <http://purl.org/swan/1.2/swan-commons/> .
@prefix swanqs: <http://purl.org/swan/1.2/qualifiers/> .
@prefix swandr: <http://purl.org/swan/1.2/discourse-relationships/> .
@prefix swanpav: <http://purl.org/swan/1.2/pav/> .
@prefix swanci: <http://purl.org/swan/1.2/citations/> .
@prefix swanag: <http://purl.org/swan/1.2/agents/> .
@prefix owl: <http://www.w3.org/2002/07/owl#> .
@prefix foaf: <http://xmlns.com/foaf/0.1/> .
<http://hypothesis.alzforum.org/researchstatement/99> a swande:ResearchStatement ;
swande:title "Tau-linked disease processes drive the onset and progression of AD and
related tauopathies"@en;
swande:description "This hypothesis suggests that tau mediates neurodegeneration. The
authors provide analysis of the progress made towards a mechanistic understanding
of tau-mediated neurodegeneration, and a discussion of the therapeutic strategies
that target the most severe toxic consequences of tau pathologies. This hypothesis
summarizes the current understanding of normal tau functions and the pathogenic
effects of tau aggregates in AD and related neurodegenerative tauopathies, in the
onset and progression of these disorders."@en;
swande:contains <http://hypothesis.alzforum.org/researchstatement/100>,
<http://hypothesis.alzforum.org/researchstatement/101>,
<http://hypothesis.alzforum.org/researchstatement/102>,
<http://hypothesis.alzforum.org/researchstatement/103>;
swanco:researchStatementQualifiedAs
<http://swan.mindinformatics.org/ontologies/1.2/rsqualifiers/hypothesis>;
swanci:derivedFrom <http://hypothesis.alzforum.org/citation/320>;
swanpav:authoredBy <http://hypothesis.alzforum.org/personname/23>,
<http://hypothesis.alzforum.org/personname/24>,
<http://hypothesis.alzforum.org/personname/25>;
swanpav:curatedBy <http://hypothesis.alzforum.org/people/gwen_wong>;
swanpav:createdBy <http://hypothesis.alzforum.org/people/elizabeth_wu>;
swanpav:createdOn "April 1, 2009".
<http://hypothesis.alzforum.org/personname/23> a swanag:PersonName;
swanag:firstName "Carlo";
swanag:lastName "Ballatore".
<http://hypothesis.alzforum.org/personname/24> a swanag:PersonName;
swanag:firstName "Virginia";
swanag:lastName "Lee".
<http://hypothesis.alzforum.org/personname/25> a swanag:PersonName;
swanag:firstName "John";
swanag:lastName "Trojanowski".
<http://hypothesis.alzforum.org/citation/320> a swanci:JournalArticle;
swanci:title "Tau-mediated neurodegeneration in Alzheimer's disease and related disorders";
swanci:contributionAuthor <http://hypothesis.alzforum.org/personname/23>,
<http://hypothesis.alzforum.org/personname/24>,
<http://hypothesis.alzforum.org/personname/25>;
swanci:contributionPublicationEnvironment <urn:issn:19403429>;
swanci:contributionPublisher <http://www.hsci.harvard.edu/>;
swanci:contributionPublishingDate "2007 Sep";
swanci:volume "8";
swanci:issue "9";
swanci:pagination "663-72";
swanci:doi "10.1038/nrn2194";
swanpav:importedOn "2009-01-24T00:00:00+05:00";
swanpav:importedFromSource <http://www.ncbi.nlm.nih.gov/pubmed/>;
swanpav:importedWithId "17684513".
<urn:issn:15320480> a swanci:Journal;
rdfs:label "Nature reviews. Neuroscience";
swanci:title "Nature reviews. Neuroscience";
swanci:shortTitle "Nat Rev Neurosci";
swanci:issnPrint "1471003X";
swanci:issnElectronic "14710048";
swanci:publishedBy <http://www.elsevier.com/>;
swanpav:importedOn "2009-01-24T00:00:00+05:00";
swanpav:importedFromSource <http://www.ncbi.nlm.nih.gov/pubmed/>.
<urn:issn:1471003X> a swanci:Journal;
rdfs:label "Nature reviews. Neuroscience";
owl:sameAs <urn:issn:14710048>;
swanpav:importedOn "2009-01-24T00:00:00+05:00";
swanpav:importedFromSource <http://www.ncbi.nlm.nih.gov/pubmed/>.
<http://hypothesis.alzforum.org/organization/32> a swanci:Publisher;
rdfs:label "Nature Pub. Group";
foaf:name "Nature Pulishing Group";
foaf:homepage "http://www.nature.com/";
swanci:place "United States";
swanpav:importedOn "2009-01-24T00:00:00+05:00";
swanpav:importedFromSource <http://www.ncbi.nlm.nih.gov/pubmed/>.
6 Usage of SKOS
SKOS (Simple Knowledge Organization System) [SKOS] provides a model for expressing the basic structure and content of concept schemes such as thesauri, classification schemes, subject heading lists, taxonomies and other similar types of controlled vocabulary. In the SWAN ecosystem, SKOS has been used to define classification/annotation through controlled vocabularies or terminologies - we call the terms belonging to the controlled vocabularies: qualifiers.
The aim of SWAN is to publish scientifically valuable curated content, the taxonomical approach is therefore implemented at the community level to solve tagging issues such as: ambiguity (a tag can have two different meanings, e.g. "apple"), heterogeneity (e.g. "apple", "pomme", "manzana") as well as the lack of organization between tags (e.g. there is no way, unless applying clustering methods, to find a link between the tag "apple" and the tag "fruit"). Ideally, the annotation should be performed through ontologies and shared knowledge bases. This is the case of genes and proteins that are expressed through the SWAN life science entities module. However, when it is not possible to reuse an existing ontology and when the development of a new ontology is not justified, we make use another mechanism based on classification/annotation through controlled vocabularies or terminologies. We call the terms belonging to the controlled vocabularies: qualifiers. They include definitions, are organized - mainly in multiple taxonomies - and can be defined for personal or public use.
In fact, in our experience, creating, documenting, publishing and maintaining OWL ontologies or RDF Schema vocabularies can be a complex and time consuming task. It is not only a mere problem of using the right tools, the bodies of knowledge to be represented are often heterogeneous, incomplete and with not well defined boundaries. This leads to the need of highly skilled and experienced knowledge engineers that should be able to leverage the usage of the proper foundation ontologies and the reuse of suitable existing domain vocabularies/ontologies. In those cases where timing is critical and little inference is required, the option of lightweight or agile classification/annotation scheme development methods can be attractive.
Example of definition of taxonomy with SKOS in RDF/XML format:
...
<skos:ConceptScheme rdf:about="&stemcellcheatsheet;nature_report_stem_cell_cheat_sheet">
<rdfs:label>Nature Report Stem Cell Cheat Sheet</rdfs:label>
<dc:title>Nature Report Stem Cell Cheat Sheet</dc:title>
<foaf:maker rdf:resource="http://www.hcklab.org/people/pc/"/>
<dc:publisher rdf:resource="http://swan.mindinformatics.org/"/>
<dc:contributor rdf:resource="http://www.hcklab.org/people/pc/"/>
<dc:contributor rdf:resource="http://swan.mindinformatics.org/people/elizabeth-wu/"/>
<swanpav:curatedBy rdf:resource="http://www.hcklab.org/people/pc/"/>
<swanpav:curatedBy rdf:resource="http://swan.mindinformatics.org/people/elizabeth-wu/"/>
<dc:creator>
<foaf:Person rdf:about="http://www.hcklab.org/people/pc/">
<foaf:name>Paolo Ciccarese</foaf:name>
</foaf:Person>
</dc:creator>
<dc:created>2009-01-24T04:35:00+05:00</dc:created>
<dcterms:issued>2009-01-25T10:00:00+05:00</dcterms:issued>
<dcterms:modified>2009-01-26</dcterms:modified>
<swanpav:lastUpdateBy rdf:resource="http://www.hcklab.org/people/pc/"/>
<swanpav:versionNumber>002</pav:versionNumber>
<skos:hasTopConcept rdf:resource="&stemcellcheatsheet;cell"/>
</skos:ConceptScheme>
<skos:Concept rdf:about="&stemcellcheatsheet;hsc">
<skos:prefLabel>HSC</skos:prefLabel>
<skos:definition>Haematopoietic stem cells (blood-forming stem cells that reside in bone marrow)</skos:definition>
<skos:broader rdf:resource="&stemcellcheatsheet;cell"/>
</skos:Concept>
...
Example of qualifier usage by a Research Statement:
...
<http://hypothesis.alzforum.org/researchstatement/989> a swande:ResearchStatement ;
swanq:qualifiedBy <&stemcellcheatsheet;hsc>;
7 Alignment between the SWAN and SIOC ontologies
Starting from version 1.2, the SWAN ontology has been aligned with the SIOC (Semantically-Interlinked Online Communities) ontology. SWAN and SIOC act in a complementary way: SWAN provides fine-grained modeling of scientific discourse elements while SIOC can represent more generic contributions of online communities. The details of the SWAN/SIOC integration are the subject of another HCLS note [LINK SWAN-SIOC].
8 Future Directions
The SWAN ontology was created to cover the requirements of a single project and along the way became the point of reference for other online communities. This trend increased the amount of effort in terms of domain ontologies we need to integrate in our ecosystem to serve multiple needs. As in the biomedical field, many domain ontologies are under intense development, we believe the SWAN ontology will become more focused on the scientific discourse providing the glue and a set of recommendations to integrate the required domain ontologies.
In addition, in order to provide better integration of existing data, besides the maping with SIOC, other mappings between existing RDFS/OWL models for argumentative discussion may be considered. Examples are IBIS [IBIS], aTags [ATAGS], SALT [SALT] and [SchoolOnto].
Appendix
A References
- [ENCY-PHIL]
- Concept of Knowledge
Klein PD - Craig E, Editor,
The Routledge Shorter Encyclopedia of Philosophy. Abingdon, Oxfordshire, UK: Routledge; 2005, p. 525.
- [WORKING-KNOWLEDGE]
- Working Knowledge
Davenport T, Prusak L.
Harvard Business School Press. Boston; 1984.
- [SWAN-ONT-JBI]
- The SWAN Biomedical Discourse Ontology
Ciccarese P, Wu E, Kinoshita J, Wong G, Ocana M, Ruttenberg A, Clark T
J Biomed Inform. 2008 Oct;41(5):739-51. Epub 2008 May 4.
- [SWAN-ONT-WEB]
- The SWAN Ontology Ecosystem , http://swan.mindinformatics.org/ontology.html .
- [OWL]
- OWL Web Ontology Language Overview,
Deborah L. McGuinness and Frank van Harmelen, Editors,
W3C Recommendation, 10 February 2004,
http://www.w3.org/TR/2004/REC-owl-features-20040210/ .
Latest version available at http://www.w3.org/TR/owl-features/ .
- [SWAN-PROJECT]
- The SWAN Project
Alzheimer Research Forum, Massachusetts General Hospital and Harvard Medical School
http://swan.mindinformatics.org/ .
- [SWAN-ALZHEIMER]
- SWAN Alzhemier Knowledge Base
Alzheimer Research Forum, Massachusetts General Hospital and Harvard Medical School
http://hypothesis.alzforum.org/ .
- [SCF]
- Science Collaboration Framework
Initiative in Innovative Computing at Harvard University
http://sciencecollaboration.org/ .
- [RDFS]
- RDF Vocabulary Description Language 1.0: RDF Schema
Dan Brickley and R.V. Guha, Editors.
W3C Recommendation, 10 February 2004,
http://www.w3.org/TR/2004/REC-rdf-schema-20040210/ .
Latest version available at http://www.w3.org/TR/rdf-schema/ .
- [FOAF]
- FOAF Vocabulary, http://xmlns.com/foaf/spec/ .
- [DC]
- Dublin Core Metadata Element Set, Version 1.1, http://dublincore.org/documents/dces/ .
- [DC-TERMS]
- Dublin Core Metadata Initiative Metadata Terms, http://dublincore.org/documents/dcmi-terms/ .
- [SKOS]
- SKOS Simple Knowledge Organization System
Alistair Miles and Sean Bechhofer, Editors
www.w3.org/TR/2009/CR-skos-reference-20090317/
Latest version available at http://www.w3.org/TR/skos-reference .
- [BIBO]
- Bibliographic Ontology
Frédérick Giasson, Editor
Available at http://bibliographicontology.com/ .
- [OWL-ED2006]
- The SWAN Biomedical Discourse Ontology
Ciccarese P, Wu E, Kinoshita J, Wong G, Ocana M, Ruttenberg A, Clark T
J Biomed Inform. 2008 Oct;41(5):739-51. Epub 2008 May 4. - [ATAGS]
-
Matthias Samwald.
aTags: Associative Tags.
http://hcls.deri.org/atag/
- [IBIS]
-
Danny Ayers.
IBIS Vocabulary: Issue-Based Information Systems for the Semantic Web.
http://hyperdata.org/xmlns/ibis/
- [SALT]
-
Tudor Groza, Siegfried Handschuh, Knud Möller and Stefan Decker.
SALT - Semantically Annotated LaTeX for Scientific Publications.
Proceedings of the 4th European Semantic Web Conference (ESWC 2007).
http://salt.semanticauthoring.org/
- [SchoolOnto]
-
The SchoolOnto project.
http://projects.kmi.open.ac.uk/scholonto/
B Acknowledgements
The ontology ecosystem presented here is the result of use cases analyses and discussions involving all members of the SWAN project team: Tim Clark, Paolo Ciccarese, June Kinoshita, Marco Ocana, Gwen Wong and Elizabeth Wu. We also would like to thank Alan Ruttenberg and Jonathan Rees of Science Commons for contributing valuable comments during the process of creation of the ontology.
We thank Alexandre Passant, Susie Stephens and David Shotton for their valuable comments and feedback during the writing and editing of this note. We also thank the entire Scientific Discourse Task Force, and more generally the Health Care and Life Science Interest Group within the W3C.



