In Oct 2004, The W3C Workshop on Semantic Web for Life Sciences brought together key leaders from Life Sciences companies, pharamceuticals, research organizations, and academics to discuss how Semantic Web technologies can help to manage modern life sciences research, enable disease understanding, and accelerate the development of therapies. The report from the conference is available and contains a number of descriptions of ongoing work.
Many participants of the Workshop who are now implementing semantic web technologies to solve life science problems, or who are considering options for doing so, expressed interest in an ongoing forum for sharing implementation experience. The public-semweb-lifesci list provides a forum for these discussions. This following is designed to provide a sample of some of the use cases, tools, applications and implementation experiences discussed on this forum to help understand the benefits Semantic Web technologies in Life Sciences. If you have example slides, or data, about example applications which will help others understand this field, and which you would like us to list here, please contact Eric Miller, em@w3.org.
Partners Healthcare System's Clinical Knowledge Management Group, a division of Clinical Informatics Research and Development, was formed in order to implement content management infrastructure and support the management of the vast amount of knowledge encoded in clinical systems across the enterprise. These ontologies and rules are served up through applications and services to support guided observation capture, guided ordering, and guided interpretation of clinical data. Workflow portals leveraging this knowledge include the electronic health record for care-givers and consumers, quality performance management, and clinical research. During phase 1, an inventory of encoded knowledge assets was performed and a meta-knowledge document library of the knowledge specifications for encoded knowledge was published to an internally developed portal. During phase 2, Documentum eRoom and Content Management Server solutions were implemented to support virtual, collaborative updating of decision support content as well as robust life-cycle management at the 'meta-knowledge" level.
This year, phase 3, we begin a series of projects to implement new knowledge-encoding editors for ontologies (Health Language Inc.; Cerebra) and rules (ILOG) which we will integrate with Documentum to support life-cycle management of in-production encoded content, inheritance/propagation of content across dependent knowledge bases, and better visualization of content-interrelationships for knowledge editors and subject matter experts alike. For example, when the definition of a contraindication to a drug such as a beta-blocker must change, this definition must be propagated to all dependent rules, order sets, documentation templates, and quality management reporting systems.
Health Language Inc. supplies medical terminologies and their inter-mappings. Cerebra supplies OWL-based ontology editors, a description-logic engine, and run-time recognition services. ILOG supplies rule editors and rule execution services. Documentum supplies collaboration, content-lifecycle management and business process management services for the sharing, cataloguing, and publishing of content.
Partners Healthcare System recognizes that the advent of personalized medicine will exponentially increase the rate of change of clinical knowledge that drives research and clinical care. This emerging knowledge management infrastructure is mission critical to reduce the cost and increase the speed of knowledge discovery and knowledge acquisition by our clinical decision support systems. The innovation adoption curve for healthcare greatly depends on a robust knowledge management infrastructure for translational medicine.
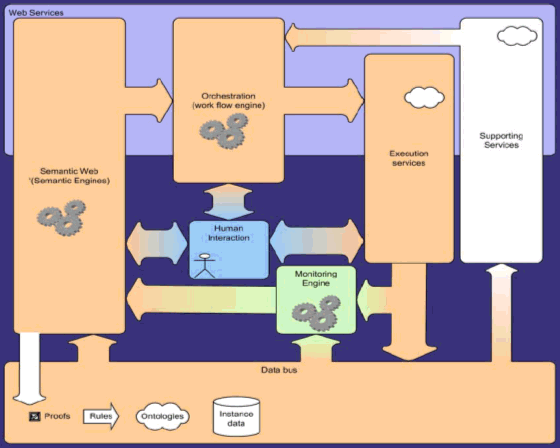
'Connected Knowledge' is the key word. If we define knowledge as a higher form of information, based on uniquely defined linked concepts and we then we can easily see the Semantic Web as a way of implementing 'connected knowledge'. As a healthcare IT vendor Agfa recognizes the high importance of the Semantic Web in achieving our goal: making systems that cross the borders of departments, hospitals, home, universities en governments and give all stakeholders intelligent tools, using knowledge and guidelines, to obtain a higher quality and more cost effective health care. Currently we focus on a Decision Support System, going beyond classical Clinical Pathways in that it dynamically creates a path, based on the current and constantly changing clinical information of the patient, the environment and the guidelines. Execution of procedures creates new data that will be taken into account to calculate the next steps in the path. We believe that one system cannot contain all medical knowledge. Therefore we make use of the fact that ontologies can be merged and connected to leverage existing and fragmented but highly specific pieces of knowledge. Dr. Dirk Colaert MD, Advanced Clinical Application Manager, Agfa HealthCare "Desirable Features of Rule Based Systems for Medical Knowledge", Position paper for W3C workshop Rule languages for Interoperability, 2005. "Towards Adaptable Clinical Pathway Using Semantic Web", Position paper for W3C workshop Semantic Web for Life Science, 2004. |
 |
The goals of Active Semantic Documents (ASD) are to reduce medical errors, improve physician efficiency and improve patient safety and satisfaction in medical practice. Semantic Web technology helps achieve these goals in an ontology driven process that involves multiple populated ontologies, automatic semantic annotation of documents, and rule processing.
ASD are documents (typically in XML based format). ASDs are semantic since they are semantically annotated using one or more relevant OWL ontologies which provide the nomenclature and conceptual model for interpreting and reasoning with the concept, and optionally annotated using lexically significant concepts and phrases (hence providing weaker semantics than the concepts and phrases that are annotated with and interpreted with respect to ontologies). ASDs are active because they support automatic and dynamic validation and decision making on the content of the document. This is accomplished typically by executing rules (such as SWRL or in the form of RDQL (with current plans of migrating this to SPARQL)) on semantic annotations and relationships that span across ontologies. Examples of semantic rule include prevention of drug interaction (i.e., not allowing a patient to be prescribed two interacting drugs) or ensuring the procedure performed has a supporting diagnoses. ASDs display the semantic and lexical annotations in document displaced in a browser, show results of rule execution, and provide the ability to modify semantic and lexical components of its content in an ontology-supported and otherwise constrained manner (such as through lists, bags of terms, specialized reference sources, or a thesaurus or lexical reference system such as WordNet). This functionality is time saving when if come to fixing broken rules due to the ability of the ASD to offer practical suggestions resolving the problem.
Active Semantic Electronic Medical Record (ASEMR) application exemplified a practical implementation of ASDs [See example with explanations]. See the ASEMR description and demo, resulting from collaboration between Athens Hearth Center (AHC) and the Large Scale Distributed Information Systems (LSDIS) lab at the University of Georgia.
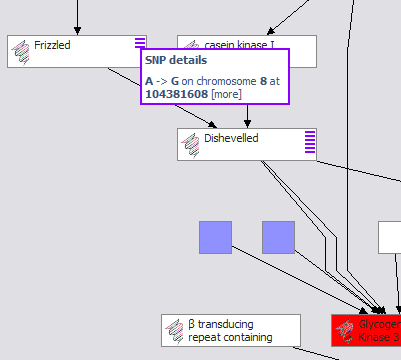
BioDASH is a Semantic Web prototype of a Drug Development Dashboard that associates disease, compounds, drug progression stages, molecular biology, and pathway knowledge for a team of users. Since such relevant information usually resides across many intranet database servers and different R&D groups, the challenge is more about leveraging the information one already has in a semantically coherent fashion than about making new data models. Here Haystack, a generic RDF-based user interface tool, is used to create "lenses" for aggregating and viewing drug and molecular pathways information used in drug discovery. Data from several sources are integrated, on the client, and explored in a variety of views. A more detailed explanation and demonstration of these tools BioDash Demo home page Thanks to Melissa Cline (Affymetrix), Joanne Luciano (BioPAX/Partners HealthCare), Eric Neumann (HCLSIG co-chair), Eric Prudhommeaux (W3C), Dennis Quan (IBM), Susie Stephens (Oracle), John Wilbanks (Science Commons), Ian Wilson (University of Colorado) for making this demonstration possible. |
 |
Life science is, at root, a collaborative discipline. Each scientist draws upon the canon of knowledge, established through experimentation and verified through peer review and publication. But modern technological approaches to managing data and papers in the life sciences in many cases can make the discovery process harder. Data is stored in different formats, exposed in different interfaces, and knowledge is locked up in document formats that don't bring the full power of computation to bear on behalf of the individual scientist. In many ways, the researcher is forced to play a complex strategy without a map. The Simile project seeks to enhance inter-operability among digital assets, schemata/vocabularies/ontologies, metadata, and services by using Semantic Web technologies. A key challenge is that the collections which must inter-operate are often distributed across individual, community, and institutional stores. While the domain focus of the Simile project is in Digitial Libraries, the problem is a very similar one to many other domains. Applying the technologies developed by the Simile project in the area of Life Sciences provides an example of how Semantic Web technologies may aid individual researchers by enabling more effective ways of data integration, management and collaboration on the Web. |
 |
Analyzing and interpreting vast repositories of scientific literature and other sources of biomedical knowledge is a daunting task to researchers in the life sciences. Language and Computing has created a powerful knowledge discovery system that offers significant value to these challenging endeavors.
This highly automated system gleans information from thousands of scientific documents, domain-specific taxonomies, public and proprietary databases and other external ontologies combining these into an integrated knowledge base. Powerful inference tools then generate concepts and relationships that can be represented visually. These relationships range from simple cause-and-effect triples to complex, transitive 3rd and 4th order relationships that would be otherwise difficult to detect. Investigators at a major pharmaceutical company are now able to explore relationships among compounds, tissue types, and body systems. This automated approach has enabled rapid explanation of in-vivo experimental results and dramatically reduced drug development time.
This new approach is currently being extended to incorporate clinical trials documents, enabling researchers to perform bedside-to- analysis that closes the loop on the behaviors of these compounds, tissue types and body systems.
Eric Miller <em@w3.org>, (W3C) Semantic Web Activity Lead