Life science is, at root, a collaborative discipline. Each scientist draws upon the canon of knowledge, established through experimentation and verified through peer review and publication. But modern technological approaches to managing data and papers in the life sciences in many cases can make the discovery process harder. Data is stored in different formats, exposed in different interfaces, and knowledge is locked up in document formats that don't bring the full power of computation to bear on behalf of the individual scientist. In many ways, the researcher is forced to play a complex strategy without a map.
The Simile project seeks to enhance inter-operability among digital assets, schemata/vocabularies/ontologies, metadata, and services by using Semantic Web technologies. A key challenge is that the collections which must inter-operate are often distributed across individual, community, and institutional stores. While the domain focus of the Simile project is in Digitial Libraries, the problem is a very similar one to many other domains.
Applying the technologies developed by the Simile project in the area of Life Sciences provides an example of how Semantic Web technologies may aid individual researchers by enabling more effective ways of data integration, management and collaboration on the Web. The following is a demonstration of these technologies using the data provided by the BioDash example.
The demonstration focuses on an investigation into the therapeutic value of GSK3beta (glycogen synthase kinase 3 beta), a regulatory enzyme associated with multiple diseases, including diabetes type 2 as well as Alzheimer’s. More detail about this scenario can be found on the BioDash home page. An important aspect to mention is the fact that this scenario simply reuses the exact same data, but provides a different way of navigating, visualizing and reusing this data.
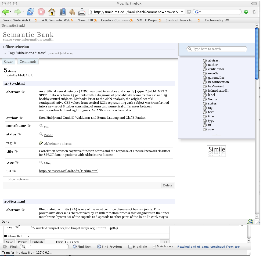
After exploring this information, a user can associate informal ad-hoc keywords (aka 'tags') or controlled vocabularies associated with any object. This act stores this information in an individuals personal data storage environment (aka "Piggy-Bank").
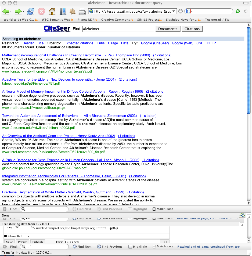
Data associated with research comes from many sources but helping contextualize how things connect is critical. The following example shows how bibliographic information from Citeseer or Pubmed might similarly be browsed, stored, organized and contextualized with other Life Sciences data.
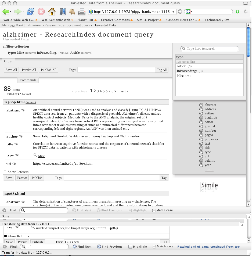
While its important for enable individual researchers to manage this information, often times there is so much data from so many areas its more effective to do this as a research team or community. When this is the case, the information can be then easily published into a collaborative Semantic Bank where others might be able to further put in context this work with other data sets, genes, proteins, hypotheses, etc..