Warning:
This wiki has been archived and is now read-only.
Draft of Use Cases
Contents
- 1 Expressivity
- 2 Migration Use Case
- 3 HCLS RDB2RDF Use Cases
- 4 BioPAX Use Cases
- 5 Integrating Enterprise Relational databases for tax control
- 6 Web applications - Wordpress
- 7 Uniprot
- 8 rCAD: RNA Comparative Analysis Database
- 9 Exposing many-to-many join tables as simple triples
- 10 Value-based type specification
Expressivity
Analysis of these use cases yields a set of functional requirements for the transformation from relational to RDF. Relational data, with it's foreign and primary keys, is a potentially cyclic graph, as is RDF. Edges in this relational graph are realised join constraints in SQL queries, often corresponding to foreign/primary key declarations in the relational schema. These edges can be expressed as RDF triples, as can all of the attribute in an relation.
Some use cases require that a relational graph be represented directly as an isomorphic RDF graph, while others require transformations of the graph. Below, the former use cases are labeled isomorphic.
example patient database graph:
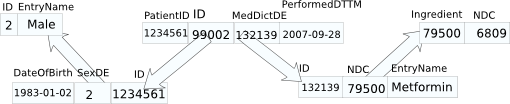
RDF graph isomorphic to patient database graph:

graphs not isomorphic to DB graph:
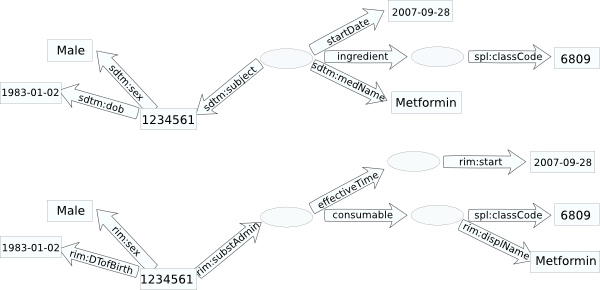
RDF identifiers for objects in the [conceptual model] can, in some cases, be generated from a transformation of the schema and data in a tuple representing that conceptual model. For example, it may be sufficient to identify a patient in a clinical database with primary key patientID and value 1234561 as http://myclinic.example/patient/patientID.12334561#x, while but if the patient IDs are shared with another database, it will be necessary to transform one or both of these identifiers into a common form, e.g. http://allclinics.example/sharedRecords/patient.12334561#y. These use cases are labeled custom-identifier.
Migration Use Case
One of the reasons for open standards is to allow easy migration between different systems. Just as a single web-page in HTML can be viewed both my two different web browsers from different vendors, a single relational data to RDF mapping standard should allow a custodian of one database to expose their data as RDF and then, when they export their data to another database, allow the newly imported data to be queried as RDF without changing the mapping file.
For example, imagine that a database administrator is working on exposing weather data as linked data to be found and consumed by other applications. At first, this weather data is stored in a light-weight database (such as MySQL). However, as more and more weather data is collected, and more and more users access this RDF, the light-weight database has difficulty scaling, providing problems for users. Therefore, the database administrator migrates their database to a more heavy-weight database (such as Oracle Database 11g).
Of course, the database administrator does not want to have re-create the ability to view the data as RDF using a vendor-specific mapping file, but instead wants to seamlessly migrate the view of their data as RDF. A standardized mapping between relational data and RDF allows the database administrator to migrate the view of their data as RDF across databases, allowing the vendors to compete on functionality and features rather than forcing database administrators to rewrite their entire relational data to RDF mapping when they want to migrate their data from one database to another.
HCLS RDB2RDF Use Cases
The Semantic Web for Health Care and Life Sciences Interest Group has created several demonstrators using SPARQL to query clinical and biological relational databases. Included is the database structure, sample data, a SPARQL query with results, and an equivalent SQL query.
COI Patient Recruitment Demo
Diabetic patient data from the University of Texas Health Science Center is expressed as RDF views corresponding to HL7/RIM and CDISK SDTM data structures. Some columns have been omitted from this use case for brevity.
While there are many motivations for providing a common interface to administratively distinct databases (access to patient history, shared rules for clinical decision support, etc), the example queries following the table description are used to find candidates for clinical studies.
Namespace Prefixes
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> . @prefix hl7: <http://www.hl7.org/v3ballot/xml/infrastructure/vocabulary/vocabulary#> . @prefix spl: <http://www.hl7.org/v3ballot/xml/infrastructure/vocabulary/vocabulary#> .
Person
| ID | MiddleName | SexDE | DateOfBirth | LastEditedDTTM | |
| 1234561 | 2 | 1983-01-02 00:00:00 | 2007-11-13 15:49:20 | ||
| 1234562 | 3 | 1963-12-27 00:00:00 | 2008-01-30 17:08:42 | ||
| 1234563 | 2 | 1983-02-25 00:00:00 | 2007-03-10 06:01:55 |
<http://hospital.example/DB/Person/ID.1234561#record> a hl7:Person ; hl7:entityName "" ; hl7:administrativeGenderCodePrintName Sex_DE:M ; hl7:livingSubjectBirthTime "1983-01-02T00:00:00Z"^^xsd:dateTime . <http://hospital.example/DB/Person/ID.1234562#record> a hl7:Person ; hl7:entityName "" ; hl7:administrativeGenderCodePrintName Sex_DE:F ; hl7:livingSubjectBirthTime "1963-12-27T00:00:00Z"^^xsd:dateTime . <http://hospital.example/DB/Person/ID.1234563#record> a hl7:Person ; hl7:entityName "" ; hl7:administrativeGenderCodePrintName Sex_DE:M ; hl7:livingSubjectBirthTime "1983-02-25T00:00:00Z"^^xsd:dateTime .
<http://hospital.example/DB/Person/ID.1234561#record> a sdtm:Patient ; stdm:middleName "" ; stdm:dateTimeOfBirth "1983-01-02T00:00:00Z"^^xsd:dateTime . <http://hospital.example/DB/Person/ID.1234562#record> a sdtm:Patient ; sdtm:middleName "" ; sdtm:dateTimeOfBirth "1963-12-27T00:00:00Z"^^xsd:dateTime ; sdtm:sex <http://hospital.example/DB/Sex_DE#F> . <http://hospital.example/DB/Person/ID.1234563#record> a sdtm:Patient ; sdtm:middleName "" ; sdtm:dateTimeOfBirth "1983-02-25T00:00:00Z"^^xsd:dateTime ; sdtm:sex <http://hospital.example/DB/Sex_DE#F> .
Sex_DE
| ID | EntryCode | EntryName | EntryMnemonic | |
| 2 | 1 | Male | M | |
| 3 | 2 | Female | F |
<http://hospital.example/DB/Zex_DE/ID.2#record>
hl7:administrativeGenderCodePrintName "Male"@en-us ;
Sex_DE:EntryMnemonic "M"@en-us .
<http://hospital.example/DB/Zex_DE/ID.3#record>
hl7:administrativeGenderCodePrintName "Female"@en-us ;
Sex_DE:EntryMnemonic "F"@en-us .
<http://hospital.example/DB/Person/ID.1234561#record>
stdm:sex <http://hospital.example/DB/Sex_DE#M> .
<http://hospital.example/DB/Person/ID.1234562#record>
stdm:sex <http://hospital.example/DB/Sex_DE#F> .
<http://hospital.example/DB/Person/ID.1234563#record>
stdm:sex <http://hospital.example/DB/Sex_DE#M> .
Item_Medication
| ID | PatientID | ItemType | PerformedDTTM | |
| 99999999002 | 1234561 | ME | 2007-09-28 00:00:00 | |
| 99999999003 | 1234562 | ME | 2007-09-28 00:00:00 | |
| 99999999004 | 1234562 | ME | 2008-07-28 00:00:00 |
<http://hospital.example/DB/Person/ID.1234561#record>
hl7:substanceAdministration <http://hospital.example/DB/Item_Medication/ID.99999999002#record> .
<http://hospital.example/DB/Item_Medication/ID.99999999002#record>
a hl7:SubstanceAdministration ;
hl7:effectiveTime _:t1 .
_:t1 hl7:start "2007-09-28T00:00:00"^^xsd:dateTime .
<http://hospital.example/DB/Person/ID.1234562#record>
hl7:substanceAdministration <http://hospital.example/DB/Item_Medication/ID.99999999003#record> ;
hl7:substanceAdministration <http://hospital.example/DB/Item_Medication/ID.99999999004#record> .
<http://hospital.example/DB/Item_Medication/ID.99999999003#record>
a hl7:SubstanceAdministration ;
hl7:effectiveTime _:t2 .
_:t2 hl7:start "2007-09-28T00:00:00"^^xsd:dateTime .
<http://hospital.example/DB/Item_Medication/ID.99999999004#record>
a hl7:SubstanceAdministration ;
hl7:effectiveTime _:t3 .
_:t3 hl7:start "2008-07-28T00:00:00"^^xsd:dateTime .
Medication
| ID | ItemID | Dose | Refill | QuantityToDispense | DaysToTake | PrescribedByID | MedDictDE | |
| 88888888002 | 99999999002 | 2 | 6 | 180 | 45 | 1004682 | 132139 | |
| 88888888003 | 99999999002 | 2 | 0 | 180 | 45 | 1004683 | 132139 | |
| 88888888004 | 99999999003 | 2 | 6 | 180 | 45 | 1004682 | 132139 | |
| 88888888005 | 99999999004 | 4 | 6 | 180 | 45 | 1004682 | 132139 |
<http://hospital.example/DB/Item_Medication/ID.99999999002#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
_:t1 hl7:durationInDays 45 .
<http://hospital.example/DB/Item_Medication/ID.99999999003#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
<http://hospital.example/DB/Item_Medication/ID.99999999004#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
_:t2 hl7:durationInDays 45 .
<http://hospital.example/DB/Item_Medication/ID.99999999005#record>
hl7:consumable <http://hospital.example/DB/Medication_DE/ID.132139#record> .
_:t3 hl7:durationInDays 45 .
Medication_DE
| ID | Entry | EntryCode | EntryName | NDC | Strength | Form | UnitOfMeasure | DrugName | DisplayName | |
| 132139 | 131933 | 98630 | GlipiZIDE-Metformin HCl 2.5-250 MG Tablet | 54868079500 | 2.5-250 | TABS | MG | GlipiZIDE-Metformin HCl | GlipiZIDE-Metformin HCl 2.5-250 MG Tablet |
<http://hospital.example/DB/Medication_DE/ID.132139#record>
hl7:displayName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" .
NDCcodes
| ingredient | RxCUI | labelType | name | NDC | |
| 6809 | 351273 | Clinical | Glipizide 2.5 MG / Metformin 500 MG Oral Tablet | 54868079500 |
<http://hospital.example/DB/Medication_DE/ID.132139#record>
spl:activeIngredient _:i1 .
_:i1 spl:classCode 54868079500 .
[ a sdtm:ConcomitantMedication ;
sdtm:subject <http://hospital.example/DB/Person/ID.1234561#record> ;
sdtm:standardizedMedicationName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" ;
hl7:activeIngredient [hl7:classCode 54868079500 ] ;
sdtm:startDateTimeOfMedication "2007-09-28 00:00:00"^^xsd:dateTime ] .
[ a sdtm:ConcomitantMedication ;
sdtm:subject <http://hospital.example/DB/Person/ID.1234562#record> ;
sdtm:standardizedMedicationName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" ;
hl7:activeIngredient [ hl7:classCode 54868079500 ] ;
sdtm:startDateTimeOfMedication "2007-09-28 00:00:00"^^xsd:dateTime ] .
[ a sdtm:ConcomitantMedication ;
sdtm:subject <http://hospital.example/DB/Person/ID.1234562#record> ;
sdtm:standardizedMedicationName "GlipiZIDE-Metformin HCl 2.5-250 MG Tablet" ;
hl7:activeIngredient [ hl7:classCode 54868079500 ] ;
sdtm:startDateTimeOfMedication "2008-07-28 00:00:00"^^xsd:dateTime ] .
Queries Over the RDF Graph.
The RDF graphs place the relational data into the Semantic Web. There are many ways to consume RDF data, integration with other data sources, inference according to OWL or RIF rules, browsing with a linked data browser like Bubbles or Tabulator. This patient recruitment use case can be realised as a SPARQL query over the RDF graphs. Following is a query which extracts patients taking a particular class of medication (an anticoagulant, in this example) and not another (weight loss, here).
PREFIX sdtm: <http://www.sdtm.org/vocabulary#>
PREFIX spl: <http://www.hl7.org/v3ballot/xml/infrastructure/vocabulary/vocabulary#>
SELECT ?patient ?dob ?sex # ?takes ?indicDate ?indicEnd ?contra
WHERE {
?patient a sdtm:Patient ;
sdtm:middleName ?middleName ;
sdtm:dateTimeOfBirth ?dob ;
sdtm:sex ?sex .
[ sdtm:subject ?patient ;
sdtm:standardizedMedicationName ?takes ;
spl:activeIngredient [ spl:classCode 6809 ] ;
sdtm:startDateTimeOfMedication ?indicDate
] .
OPTIONAL {
[ sdtm:subject ?patient ;
sdtm:standardizedMedicationName ?disqual ;
spl:activeIngredient [ spl:classCode 11289 ]
sdtm:startDateTimeOfMedication ?indicDate
] }
} LIMIT 30
RDF Graphs as Non-materialized Views
The RDF graphs defined by a mapping over a relational database need not be materialised. Indeed, queries or inference may be expressed in terms of a notional representation of relational data, but executed over the original relational store. Executing the following SQL query over any data in the relational tables should give results identical to a SPARQL query over the RDF graph mapped from that relational data:
SELECT R_patient.ID AS patient, R_patient.DateOfBirth AS dob, R__0_sexEntry.EntryName AS sex
FROM Person AS R_patient
INNER JOIN Sex_DE AS R_sexEntry
INNER JOIN Item_Medication AS R_indicItem
INNER JOIN Medication AS R_indicMed
INNER JOIN Medication_DE AS R_indicDE
INNER JOIN NDCcodes AS R_indicCode
LEFT OUTER JOIN (
SELECT 6 AS _DISJOINT_, R_disqualMed.MedDictDE AS disqualDE,
R_disqualItem.PerformedDTTM AS disqualDate, R_disqualItem.ID AS disqualItem,
R_disqualMed.ID AS disqualMed, R_disqualDE.NDC AS disqualNDC,
R_disqualItem.PatientID AS patient, R_disqualItem.EntryName AS takes
FROM Item_Medication AS R_disqualItem
INNER JOIN Medication AS R_disqualMed
INNER JOIN Medication_DE AS R_disqualDE
INNER JOIN NDCcodes AS R_disqualCode
WHERE (R_disqualCode.NDC=R_disqualDE.NDC)
AND (R_disqualCode.ingredient=11289)
AND (R_disqualDE.ID=R_disqualMed.MedDictDE)
AND (R_disqualItem.EntryName IS NOT NULL)
AND (R_disqualItem.PatientID IS NOT NULL)
AND (R_disqualItem.PerformedDTTM IS NOT NULL)
AND (R_disqualMed.ID IS NOT NULL)
AND (R_disqualMed.ItemID=R_disqualItem.ID)
) AS G_opt6 ON (G_opt6.patient=R_patient.ID)
AND (G_opt6.takes=R_indicItem.EntryName)
WHERE (R_indicCode.NDC=R_indicDE.NDC)
AND (R_indicCode.ingredient=6809)
AND (R_indicDE.ID=R_indicMed.MedDictDE)
AND (R_indicItem.EntryName IS NOT NULL)
AND (R_indicItem.PatientID=R_patient.ID)
AND (R_indicItem.PerformedDTTM IS NOT NULL)
AND (R_indicMed.ID IS NOT NULL)
AND (R_indicMed.ItemID=R_indicItem.ID)
AND (R_patient.DateOfBirth IS NOT NULL)
AND (R_patient.MiddleName IS NOT NULL)
AND (R_sexEntry.EntryName IS NOT NULL)
AND (R_sexEntry.ID=R_patient.SexDE);
This query is not a normative representation of the earlier SPARQL query; it is included here only to illustrate the ability of R2ML tools to produce efficient SQL queries.
BioPAX Use Cases
Set of use cases mapping pathway data to BioPAX or PRO .
DB 2 BioPAX
Namespace Prefixes
@prefix bp: <http://www.biopax.org/release/biopax-level1.owl#> . @prefix bpdemo: <biopax-short-demo-pathway.rdf#> .
Molecule Tables
Molecules
| _pk | NAME | SHORTNAME | MOLECULAR-WEIGHT | CHEMICAL-FORMULA | STRUCTURE | AVAILABILITY | |
| 10 | alpha-D-glucose | a-D-glu | 180.16 | C6H12O6 | 12 | see http://www.amaze.ulb.ac.be/ |
MoleculeSynonyms
| MOLECULE | SYNONYM | |
| 10 | α-D-glucose |
MoleculeCrossReferences
| MOLECULE | CROSSREFERENCE | |
| 10 | 11 |
MoleculePhysicalEntityParticipant
| _pk | PHYSICAL-ENTITY_molecule | STOICHIOMETRIC-COEFFICIENT | CELLULAR-LOCATION_physent | |
| 9 | 10 | 1.0 | 31 |
MoleculeDataSources
| MOLECULE | DATASOURCE | |
| 10 | 14 |
Structures
| _pk | FORMAT | DATA | |
| 12 | SMILES | C1(C(O)C(O)C(O)C(O1)CO)(O) |
Protein Tables
Protiens
| _pk | NAME | SHORTNAME | SEQUENCE | ORGANISM | AVAILABILITY | |
| 32 | glucokinase | GLK | MTK...HIL @blob@ | 33 | see http://www.amaze.ulb.ac.be/ |
ProteinSynonyms
| PROTEIN | SYNONYM | |
| 32 | Glucose kinase | |
| 32 | GLK_ECOLI |
ProteinCrossReferences
| PROTEIN | CROSSREFERENCE | |
| 32 | 35 |
ProtienPhysicalEntityParticipant
| _pk | PHYSICAL-ENTITY_protein | STOICHIOMETRIC-COEFFICIENT | CELLULAR-LOCATION_ocv | |
| 31 | 32 | 1.0 | 15 @@ |
ProteinDataSources
| PROTEIN | DATASOURCE | |
| 32 | 14 |
Other Tables
BioSources
| _pk | NAME | TAXON-XREF | |
| 33 | Escherichia coli | 34 |
UnificationCrossReferences
| _pk | ID | DB | |
| 11 | C00267 | KEGG | |
| 34 | 562 | taxon | |
| 35 | P46880 | Swiss-Prot/TrEMBL |
DataSources
| _pk | NAME | |
| 14 | aMAZE |
bpdemo:smallMolecule10 a bp:smallMolecule ;
bp:NAME "alpha-D-glucose"^^xsd:string ;
bp:SHORT-NAME "a-D-glu"^^xsd:string ;
bp:SYNONYMS "α-D-glucose"^^xsd:string ;
bp:MOLECULAR-WEIGHT 180.16 ;
bp:CHEMICAL-FORMULA "C6H12O6"^^xsd:string ;
bp:STRUCTURE bpdemo:chemicalStructure12 ;
bp:XREF bpdemo:unificationXref11 ;
bp:XREF bpdemo:unificationXref13 ;
bp:DATA-SOURCE bpdemo:KB_439584_Individual_47 ; # !! doesn't fit
bp:DATA-SOURCE bpdemo:dataSource14 ;
bp:AVAILABILITY "see http://www.amaze.ulb.ac.be/"^^xsd:string .
bpdemo:physicalEntityParticipant9 a bp:physicalEntityParticipant ;
bp:STOICHIOMETRIC-COEFFICIENT 1.0 ;
bp:PHYSICAL-ENTITY bpdemo:smallMolecule10 ;
bp:CELLULAR-LOCATION bpdemo:physicalEntityParticipant31 .
bpdemo:physicalEntityParticipant31 a bp:physicalEntityParticipant ;
bp:STOICHIOMETRIC-COEFFICIENT 1.0 ;
bp:CELLULAR-LOCATION bpdemo:openControlledVocabulary15 ;
bp:PHYSICAL-ENTITY bpdemo:protein32 .
bpdemo:openControlledVocabulary15 a openControlledVocabulary ;
bp:TERM "cytoplasm" ;
bp:XREF bpdemo:unificationXref16 .
bpdemo:protein32 a bp:protein ;
bp:NAME "glucokinase"^^xsd:string ;
bp:SHORT-NAME "GLK"^^xsd:string ;
bp:SEQUENCE "MTK...HIL"^^xsd:string ;
bp:ORGANISM bpdemo:bioSource33 .
bp:SYNONYMS "Glucose kinase"^^xsd:string ;
bp:SYNONYMS "GLK_ECOLI"^^xsd:string ;
bp:XREF bpdemo:unificationXref35 ;
bp:DATA-SOURCE bpdemo:KB_439584_Individual_48 ; # doesn't fit
bp:DATA-SOURCE bpdemo:dataSource14 ;
bp:AVAILABILITY "see http://www.amaze.ulb.ac.be/"^^xsd:string ;
bpdemo:bioSource33 a bp:bioSource ;
bp:NAME "Escherichia coli"^^xsd:string ;
bp:TAXON-XREF bpdemo:unificationXref34 .
bpdemo:unificationXref11 a bp:unificationXref ;
bp:ID "C00267"^^xsd:string ;
bp:DB "KEGG"^^xsd:string .
bpdemo:unificationXref34 a bp:unificationXref
bp:ID "562"^^xsd:string ;
bp:DB "taxon"^^xsd:string .
bpdemo:unificationXref35 a bp:unificationXref ;
bp:ID "P46880"^^xsd:string ;
bp:DB "Swiss-Prot/TrEMBL"^^xsd:string .
bpdemo:chemicalStructure12 a bp:chemicalStructure ;
bp:STRUCTURE-FORMAT "SMILES"^^xsd:string ;
bp:STRUCTURE-DATA "C1(C(O)C(O)C(O)C(O1)CO)(O)"^^xsd:string .
Integrating Enterprise Relational databases for tax control
Responsible: Angela Fogarolli
Goal: Integrating relational databases and exposing them on the web or intranet based on the final RDB2RDF XG 1.1.3 and 1.1.2 use cases ) through the use of unique identifiers. This approach consist of integrating and interlinking data about entities on different databases.
Problem:
a. Join between entities described in different databases.
b. Join structured data (SQL) to structured data, from incompatible schema, or where data is dirty, poorly normalized, lacking proper keys/indices.
Requirements:
- mapping RDB2RDF (need to write User friendly tools that implement the mappings);
- federated SPARQL queries (SPARQL2SQL...);
- unique identifiers ;
Use Case Description: This use case is a pilot project for the Trentino region tax agency. Trentino is an autonomous region in the north of Italy. The region has a population of 1 mil. People and more than 200 municipalities with their on information systems. The goal of the project is to integrate and link tax related data about people, organizations, buildings etc. This data come from different databases especially from the region’s many municipalities, each with their own individual data structures, and other sources. With our methodology we will provide a lightweight method for aggregating the data. In this way we are providing the user, a tax agent in our case an intelligent tool for navigating through the data present in the many different databases. The tool aggregates data and creates a profile for each tax payer. Each user profile shows different type of information , with links to other entities such as the buildings owned , payments made , location of residence etc.
Example
Supposed that we have two tables (Anagrafe and Urban_Cadastre) from different databases, we select some typical attributes for the two tables to explain our conversion method. Table “Angrafe” includs the information about two type of entities, persons and locations (person’s residence place), and some other information which are not properties of persons or locations:
| Firstname | Lastname | City_Residence_Place | Country_Residence_Place | Other_Info |
| Paolo | Bouquet | Trento | Italy | xyz... |
Table “Urban_Cadastre” contains the information about buildings and their owners:
| Owner_Name | Building_LocalID | Building_Address | Building_Type |
| Paolo Bouquet | 123456 | VIA G.LEOPARDI | 3 |
DDL
Using traditional RDB2RDF translation methods the RDF representation for the two example tables coming from two different databases is shown below:
<rdf:RDF xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#" xmlns:database_anagrafe="http://www.database1.org/anagrafe/"> <rdf:Description rdf:about="http://www.database1.org/anagrafe/entry_row1"> <database_anagrafe:Other_Info>xyz</database_anagrafe:Other_Info> <database_anagrafe:Country_Residence_Place>Italy</database_anagrafe:Country_Residence_Place> <database_anagrafe:City_Residence_Place>Trento</database_anagrafe:City_Residence_Place> <database_anagrafe:Last_Name>Bouquet</database_anagrafe:Last_Name> <database_anagrafe:First_Name>Paolo</database_anagrafe:First_Name> </rdf:Description> </rdf:RDF>
<rdf:RDF xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#" xmlns:database_urbano="http://www.database1.org/Urban_Cadastre/"> <rdf:Description rdf:about="http://www.database1.org/Urban_Cadastre/entry_row1"> <database_urbano:Building_Type>3</database_urbano:Building_Type> <database_urbano:Building_Address>VIA G.LEOPARDI</database_urbano:Building_Address> <database_urbano:Building_LocalID>123456</database_urbano:Building_LocalID> <database_urbano:Owner_Name>Paolo Bouquet</database_urbano:Owner_Name> </rdf:Description> </rdf:RDF>
Querying
In SQL there is no way to create a query which joins data of two tables coming from different databases. For solving the identity problem which is required by the use case creating a RDF representation is not enough. The use case demands the use of unique identifier to refer to entities in order to join descriptions about the same entity coming from different datasources.
Web applications - Wordpress
In order to make the Semantic Web useful to ordinary Web users, RDF and OWL have to be deployed on the Web on a much larger scale. Web applications such as Content Management Systems, online shops or community applications (e.g. Wikis, Blogs, Fora) already store their data in relational databases. Providing a standardized way to map the relational data structures behind these Web applications into RDF, RDF-Schema and OWL will facilitate broad penetration and enrich the Web with RDF data and ontologies and facilitate novel semantic browsing and search applications.
By supporting the long tail of Web applications and thus counteracting the centralization of the Web 2.0 applications the planned RDB2RDF standardization will help to give control over data back to end-users and thus promote a democratization of the Web.
To support this usecase scenario, the mapping language should be easily implementable for lightweight Web applications and have a shallow learning curve to foster early adoption by Web developers.
We illustrate this use case with the example of Wordpress. Wordpress is a popular blogging Web application and installed on tens of thousands of Web servers. Wordpress used a relational database (MySQL) with a relatively simple schema:
- SQL Schema
- SQL Schema and complete data of blog.aksw.org
- NTriples export of the above database created using Triplify
A mapping should be able to reuse existing vocabularies.
Mapped to RDF the resulting ontology should contain the classes post, attachment, tag, category, user and comment. An example instance of the post class, for example, should look like:
<http://blog.aksw.org/triplify/post/8> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://rdfs.org/sioc/ns#Post> . <http://blog.aksw.org/triplify/post/8> <http://rdfs.org/sioc/ns#has_creator> <http://blog.aksw.org/triplify/user/5> . <http://blog.aksw.org/triplify/post/8> <http://purl.org/dc/terms/created> "2007-02-27T17:23:36"^^<http://www.w3.org/2001/XMLSchema#dateTime> . <http://blog.aksw.org/triplify/post/8> <http://purl.org/dc/elements/1.1/title> "Submissions open: 3rd Workshop on Scripting for the Semantic Web" . <http://blog.aksw.org/triplify/post/8> <http://rdfs.org/sioc/ns#content> "The submissions web-site is now open for the 3rd Workshop on Scripting for the Semantic Web..." . <http://blog.aksw.org/triplify/post/8> <http://purl.org/dc/terms/modified> "2008-02-22T21:41:00"^^<http://www.w3.org/2001/XMLSchema#dateTime> .
An example SPARQL query on the resulting ontology could be:
SELECT ?name, ?title
WHERE {
?post rdf:type sioc:Post .
?post dc:title ?title .
?post dc:has_creator ?author
?author foaf:name ?name
}
Uniprot
+---------------------+ | Tables_in_uniProt | +---------------------+ | accToKeyword | +--------+---------+ | acc | keyword | +--------+---------+ | Q197F8 | 1 | | Q197F8 | 2 | +--------+---------+ | accToTaxon | +--------+--------+ | acc | taxon | +--------+--------+ | Q197F8 | 345201 | | Q197F7 | 345201 | +--------+--------+ | author | +----+-------------+ | id | val | +----+-------------+ | 1 | Delhon G. | | 2 | Tulman E.R. | +----+-------------+ | citation | +----+--------+-----------+----+ | id | acc | reference | rp | +----+--------+-----------+----+ | 1 | Q197F8 | 1 | 1 | | 2 | Q197F7 | 1 | 1 | +----+--------+-----------+----+ | citationRc | +----------+--------+-------+ | citation | rcType | rcVal | +----------+--------+-------+ | 101 | 1 | 1 | | 102 | 1 | 2 | +----------+--------+-------+ | citationRp | +----+------------------------------------------------+ | id | val | +----+------------------------------------------------+ | 1 | NUCLEOTIDE SEQUENCE [LARGE SCALE GENOMIC DNA]. | | 2 | GENOME REANNOTATION. | +----+------------------------------------------------+ | comment | +--------+-------------+------------+ | acc | commentType | commentVal | +--------+-------------+------------+ | Q91G88 | 1 | 1 | | Q91G88 | 1 | 2 | +--------+-------------+------------+ | commentType | +----+----------------------+ | id | val | +----+----------------------+ | 1 | SIMILARITY | | 2 | SUBCELLULAR LOCATION | +----+----------------------+ | commentVal | +----+--------------------------------------------------+ | id | val | +----+--------------------------------------------------+ | 1 | Belongs to the IIV-6 006L/238R/313L/468L family. | | 2 | Contains 1 KilA-N domain. | +----+--------------------------------------------------+ | commonName | +--------+---------------------------+ | taxon | val | +--------+---------------------------+ | 345201 | IIV-3 | | 345201 | Mosquito iridescent virus | +--------+---------------------------+ | description | +--------+---------------------------------------------+ | acc | val | +--------+---------------------------------------------+ | Q197F8 | RecName: Full=Uncharacterized protein 002R; | | Q197F7 | RecName: Full=Uncharacterized protein 003L; | +--------+---------------------------------------------+ | displayId | +--------+-----------+ | acc | val | (display ID) +--------+-----------+ | Q197F8 | 002R_IIV3 | | Q197F7 | 003L_IIV3 | +--------+-----------+ | extDb | +----+--------+ | id | val | +----+--------+ | 1 | EMBL | | 2 | RefSeq | +----+--------+ | extDbRef | +--------+-------+-------------+------------+---------+ | acc | extDb | extAcc1 | extAcc2 | extAcc3 | +--------+-------+-------------+------------+---------+ | Q197F8 | 1 | DQ643392 | ABF82032.1 | - | | Q197F8 | 2 | YP_654574.1 | - | | +--------+-------+-------------+------------+---------+ | feature | +--------+-------+-----+--------------+-------------+-------------+-----------+ | acc | start | end | featureClass | featureType | softEndBits | featureId | +--------+-------+-----+--------------+-------------+-------------+-----------+ | Q197F8 | 0 | 458 | 1 | 1 | 0 | 1 | | Q197F7 | 0 | 156 | 1 | 2 | 0 | 2 | +--------+-------+-----+--------------+-------------+-------------+-----------+ | featureClass | +----+--------+ | id | val | +----+--------+ | 1 | CHAIN | | 2 | SIGNAL | +----+--------+ | featureId | +----+----------------+ | id | val | +----+----------------+ | 1 | PRO_0000377938 | | 2 | PRO_0000377939 | +----+----------------+ | featureType | +----+------------------------------+ | id | val | +----+------------------------------+ | 1 | Uncharacterized protein 002R | | 2 | Uncharacterized protein 003L | +----+------------------------------+ | gene | +--------+-----------+ | acc | val | +--------+-----------+ | Q197F8 | IIV3-002R | | Q197F7 | IIV3-003L | +--------+-----------+ | geneLogic | +--------+---------------------+ | acc | val | +--------+---------------------+ | Q197F8 | ORFNames=IIV3-002R; | | Q197F7 | ORFNames=IIV3-003L; | +--------+---------------------+ | history | +----+---------+-------+------+-----------------------------------------------------+---------------------+--------+ | ix | startId | endId | who | what | modTime | errata | +----+---------+-------+------+-----------------------------------------------------+---------------------+--------+ | 1 | 0 | 10 | kent | New | 2009-08-23 12:31:16 | NULL | | 2 | 0 | 0 | kent | Add contents of 1 text file(s) to table varProtein. | 2009-08-23 12:31:16 | NULL | +----+---------+-------+------+-----------------------------------------------------+---------------------+--------+ | info | +--------+-----------+--------+-----------+------------+------------+------------+-----------+ | acc | isCurated | aaSize | molWeight | createDate | seqDate | annDate | organelle | +--------+-----------+--------+-----------+------------+------------+------------+-----------+ | Q197F8 | 1 | 458 | 53921 | 2009-06-16 | 2006-07-11 | 2009-07-28 | 0 | | Q197F7 | 1 | 156 | 17043 | 2009-06-16 | 2006-07-11 | 2009-07-28 | 0 | +--------+-----------+--------+-----------+------------+------------+------------+-----------+ | keyword | +----+------------------------+ | id | val | +----+------------------------+ | 1 | Complete proteome | | 2 | Virus reference strain | +----+------------------------+ | organelle | +----+--------------------------+ | id | val | (organelle name) +----+--------------------------+ | 1 | Plasmid megaplasmid Rsp. | | 2 | Plasmid pZM2. | +----+--------------------------+ | otherAcc | +--------+--------+ | acc | val | (secondary accession IDs) +--------+--------+ | P15711 | Q4N2B5 | | P68744 | P18556 | +--------+--------+ | pathogenHost | +----------+-------+ | pathogen | host | +----------+-------+ | 345201 | 7163 | | 345201 | 42431 | +----------+-------+ | protein | +--------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+ | acc | val | +--------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+ | Q197F8 | MASNTVSAQGGSNRPVRDFSNIQDVAQFLLFDPIWNEQPGSIVPWKMNREQALAERYPELQTSEPSEDYSGPVESLELLPLEIKLDIMQYLSWEQISWCKHPWLWTRWYKDNVVRVSAITFEDFQREYAFPEKIQEIHFTDTRAEEIKAILETTPNVTRLVIRRIDDMNYNTHGDLGLDDLEFLTHLMVEDACGFTDFWAPSLTHLTIKNLDMHPRWFGPVMDGIKSMQSTLKYLYIFETYGVNKPFVQWCTDNIETFYCTNSYRYENVPRPIYVWVLFQEDEWHGYRVEDNKFHRRYMYSTILHKRDTDWVENNPLKTPAQVEMYKFLLRISQLNRDGTGYESDSDPENEHFDDESFSSGEEDSSDEDDPTWAPDSDDSDWETETEEEPSVAARILEKGKLTITNLMKSLGFKPKPKKIQSIDRYFCSLDSNYNSEDEDFEYDSDSEDDDSDSEDDC | | Q197F7 | MYQAINPCPQSWYGSPQLEREIVCKMSGAPHYPNYYPVHPNALGGAWFDTSLNARSLTTTPSLTTCTPPSLAACTPPTSLGMVDSPPHINPPRRIGTLCFDFGSAKSPQRCECVASDRPSTTSNTAPDTYRLLITNSKTRKNNYGTCRLEPLTYGI | +--------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+ | proteinEvidence | +--------+---------------------+ | acc | proteinEvidenceType | +--------+---------------------+ | Q197F8 | 4 | | Q197F7 | 4 | +--------+---------------------+ | proteinEvidenceType | +----+------------------------------+ | id | val | +----+------------------------------+ | 1 | Evidence at protein level | | 2 | Evidence at transcript level | +----+------------------------------+ | rcType | +----+--------+ | id | val | +----+--------+ | 1 | STRAIN | | 2 | TISSUE | +----+--------+ | rcVal | +----+--------+ | id | val | +----+--------+ | 1 | Ankara | | 2 | Muguga | +----+--------+ | reference | +----+-------------------------------------------------------------------------------------------------------------------------------------+-------------------------------+----------+----------+------------------------+ | id | title | cite | pubMed | medline | doi | +----+-------------------------------------------------------------------------------------------------------------------------------------+-------------------------------+----------+----------+------------------------+ | 1 | Genome of invertebrate iridescent virus type 3 (mosquito iridescent virus). | J. Virol. 80:8439-8449(2006). | 16912294 | | 10.1128/JVI.00464-06 | | 2 | Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. | Virology 286:182-196(2001). | 11448171 | 21342589 | 10.1006/viro.2001.0963 | +----+-------------------------------------------------------------------------------------------------------------------------------------+-------------------------------+----------+----------+------------------------+ | referenceAuthors | +-----------+--------+ | reference | author | +-----------+--------+ | 1 | 1 | | 1 | 2 | +-----------+--------+ | tableDescriptions | | taxon | +--------+---------------------------------+----------------------------------------------------------------------+ | id | binomial | toGenus | +--------+---------------------------------+----------------------------------------------------------------------+ | 345201 | Invertebrate iridescent virus 3 | Viruses; dsDNA viruses, no RNA stage; Iridoviridae; Chloriridovirus. | | 176652 | Invertebrate iridescent virus 6 | Viruses; dsDNA viruses, no RNA stage; Iridoviridae; Iridovirus. | +--------+---------------------------------+----------------------------------------------------------------------+ | varAcc | +----------+--------+---------+ | varAcc | parAcc | variant | +----------+--------+---------+ | P15455-2 | P15455 | 2 | | P48347-2 | P48347 | 2 | +----------+--------+---------+ | varProtein | +----------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+ | acc | val | +----------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+ | P15455-2 | MGKVIPGCAETFQDSSEFQPRFEGQGQSQRFRDMHQKVEHIRSGDTIATTPGVAQWFYNDGQEPLVIVSVFDLASHQNQLDRNPRPFYLAGNNPQGQVWLQGREQQPQKNIFNGFGPEVIAQALKIDLQTAQQLQNQDDNRGNIVRVQGPFGVIRPPLRGQRPQEEEEEEGRHGRHGNGLEETICSARCTDNLDDPSRADVYKPQLGYISTLNSYDLPILRFIRLSALRGSIRQNAMVLPQWNANANAILYVTDGEAQIQIVNDNGNRVFDGQVSQGQLIAVPQGFSVVKRATSNRFQWVEFKTNANAQINTLAGRTSVLRGLPLEVITNGFQISPEEARRVKFNTLETTLTHSSGPASYGRPRVAAA | | P48347-2 | MENEREKQVYLAKLSEQTERYDEMVEAMKKVAQLDVELTVEERNLVSVGYKNVIGARRASWRILSSIEQKEESKGNDENVKRLKNYRKRVEDELAKVCNDILSVIDKHLIPSSNAVESTVFFYKMKGDYYRYLAEFSSGAERKEAADQSLEAYKAAVAAAENGLAPTHPVRLGLALNFSVFYYEILNSPESACQLAKQAFDDAIAELDSLNEESYKDSTLIMQLLRDNLTLWTSDLNEEGDERTKGADEPQDEV | +----------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+ +---------------------+
rCAD: RNA Comparative Analysis Database
Responsible: Juan Sequeda
Goal: Exposing rCAD relational databases as RDF with a SPARQL endpoint and linking it to other datasets in the life science Linked Data cloud
Requirements:
- mapping part of a schema to an existing domain ontology
- automatically generating an ontology from the SQL-DDL, because a domain ontology does not exist
rCAD - RNA Comparative Analysis using SQLServer: The tremendous increase in available biological information creates opportunities to decipher the structure, function and evolution of cellular components while presenting new computational challenges for performance and scalability. To fully utilize this large increase in knowledge, it must be organized for efficient retrieval and integrated for multi-dimensional analysis. Given this, biologists are able to invent new comparative sequence analysis protocols that will yield new and different structural and functional information. Based on Microsoft SQL-server, we have designed and implemented the RNA Comparative Analysis Database -rCAD which supports comparative analysis of RNA sequence and structure, and unites, for the first time in a single environment, multiple dimensions of information necessary for alignment viewing, sequence metadata, structural annotations, structure prediction studies, structural statistics of different motifs, and phylogenetic analysis. This system provides a queryable environment that hosts efficient updates and rich analytics.
The rCAD consists of different schema: Sequence Metadata, Evolutionary Relationships, Structural Relationships and Sequence Alignment.
For this use-case, we will be presenting only the Sequence Alignment schema. The SQL-DDL for Microsoft SQL Server can be found here: rCAD Sequence Alignment SQL DDL
This use-case presents the issue of a schema that does not have an existing domain ontology in which it can be mapped to. The closest domain ontology is the Multiple Alignment Ontology (MAO) which can only be mapped to the Sequence Alignment part of the entire rCAD database. However MAO is in the OBO language. Nevertheless, OBO ontologies can be translated to OWL (and back).
Use Case: Expose Relational Data as RDF when there is no existing Domain Ontology to map the relational schema to
Rob, from the RNA lab would like to expose the Sequence Alignment data from the rCAD database as RDF. However, there is no existing domain ontology in which the relational schema can be mapped to. Therefore, an ontology should be derived automatically from the relational schema. The RDF data will be come instance of this automatically generated ontology.
The Alignment table from the rCAD database is the following:
CREATE TABLE [AlignmentClassic].[Alignment] (
[AlnID] int NOT NULL,
[SeqTypeID] tinyint NOT NULL,
[AlignmentName] varchar(max) NULL,
[ParentAlnID] int NULL,
[NextColumnNumber] int NOT NULL,
CONSTRAINT [PK_Alignment] PRIMARY KEY([AlnID])
)
ON [PRIMARY]
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_SequenceType]
FOREIGN KEY([SeqTypeID])
REFERENCES [dbo].[SequenceType]([SeqTypeID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_Alignment]
FOREIGN KEY([ParentAlnID])
REFERENCES [AlignmentClassic].[Alignment]([AlnID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
and the desired ontology that is generated automatically from the relational schema is the following:
PREFIX rcad: <http://rcad.org/vocabulary/rcad.owl#> rcad:Alignment rdf:type owl:Class . rcad:AlignmentName rdf:type owl:DatatypeProperty. rcad:AlignmentName rdfs:domain rcad:Alignment. rcad:AlignmentName rdfs:range xsd:string. rcad:NextColumnNumber rdf:type owl:DatatypeProperty. rcad:NextColumnNumber rdfs:domain rcad:Alignment. rcad:NextColumnNumber rdfs:range xsd:integer. rcad:ParentAlnID rdf:type owl:ObjectProperty rcad:ParentAlnID rdfs:domain rcad:Alignment rcad:ParentAlnID rdfs:range rcad:Alignment
and the desired RDF triples, which are instances of the automatically generated ontology are the following:
PREFIX rcad: <http://rcad.org/vocabulary/rcad.owl#> PREFIX rcad-data: <http://rcad.org/vocabulary/rcad-data.rdf#> rcad-data:alignment1000 a rcad:Alignment; rcad:AlignmentName "My Alignment 1000"^^xsd:string; rcad:NextColumnNumber "123"^^xsd:int; rcad:ParentAlnID rcad-data:alignment2000
Use Case: Expose Mapping a Relational Database to an OWL DL ontology
Rob, from the RNA lab would like to expose the Sequence Alignment data from the rCAD database as RDF. Just recently the Multiple Alignment Ontology (MAO) as been released, which is an "ontology for data retrieval and exchange in the fields of multiple DNA/RNA alignment, protein sequence and protein structure alignment." However, this ontology has been developed in OBO. Nevertheless, OBO ontologies can be translated to OWL ontologies, specifically OWL DL. Therefore, Rob will like to map his rCAD database to the MAO ontology
The Alignment and Alignment Column tables from the rCAD database is the following:
CREATE TABLE [AlignmentClassic].[Alignment] (
[AlnID] int NOT NULL,
[SeqTypeID] tinyint NOT NULL,
[AlignmentName] varchar(max) NULL,
[ParentAlnID] int NULL,
[NextColumnNumber] int NOT NULL,
CONSTRAINT [PK_Alignment] PRIMARY KEY([AlnID])
)
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_SequenceType]
FOREIGN KEY([SeqTypeID])
REFERENCES [dbo].[SequenceType]([SeqTypeID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
ALTER TABLE [AlignmentClassic].[Alignment]
ADD CONSTRAINT [FK_Alignment_Alignment]
FOREIGN KEY([ParentAlnID])
REFERENCES [AlignmentClassic].[Alignment]([AlnID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
---
CREATE TABLE [AlignmentClassic].[AlignmentColumn] (
[AlnID] int NOT NULL,
[ColumnNumber] int NOT NULL,
[ColumnOrdinal] int NOT NULL,
CONSTRAINT [PK_AlignmentColumn] PRIMARY KEY([AlnID],[ColumnNumber])
)
GO
ALTER TABLE [AlignmentClassic].[AlignmentColumn]
ADD CONSTRAINT [FK_AlignmentColumn_Alignment]
FOREIGN KEY([AlnID])
REFERENCES [AlignmentClassic].[Alignment]([AlnID])
ON DELETE NO ACTION
ON UPDATE NO ACTION
GO
This is just part of the Multiple Alignment Ontology in OWL DL
PREFIX ro: <http://purl.org/obo/owl/RO#> PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> PREFIX owl: <http://www.w3.org/2002/07/owl#> PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> PREFIX _global: <http://purl.org/obo/owl/_global#> _global:part_of rdf:type owl:ObjectProperty _global:part_of rdf:type owl:TransitiveProperty _global:part_of rdfs:label "part_of" ro:RO_0000031 rdf:type owl:Class ro:RO_0000031 rdfs:label "sub_alignment" genid:A129876 rdf:type owl:Restriction ro:RO_0000031 rdfs:subClassOf genid:A129876 genid:A129876 owl:onProperty _global:part_of genid:A129876 owl:someValuesFrom ro:RO_0000000 ro:RO_0000084 rdf:type owl:Class ro:RO_0000084 rdfs:label "alignment_column" genid:A129906 rdf:type owl:Restriction ro:RO_0000084 rdfs:subClassOf genid:A129906 genid:A129906 owl:onProperty _global:part_of genid:A129906 owl:someValuesFrom ro:RO_0000031
Exposing many-to-many join tables as simple triples
The community's school district maintains an RDB with basic student and personnel information, including a STUDENTS and a TEACHERS table. The relationship between the two is given in a STUDENT_TEACHER table:
+------------+-------------+ | StudentId | StudentName | +------------+-------------+ | 1 | Lee | | 2 | Eric | | 3 | Juan | | ... | ... | +------------+-------------+ +------------+-------------+ | StudentId | TeacherID | +------------+-------------+ | 1 | 1 | | 1 | 2 | | 2 | 3 | | 3 | 2 | | ... | ... | +------------+-------------+ +------------+-------------+ | TeacherID | TeacherName | +------------+-------------+ | 1 | Michael | | 2 | Ahmed | | 3 | Marcelo | | ... | ... | +------------+-------------+
The SQL DDL for these tables are the following:
CREATE TABLE student ( studentID int PRIMARY KEY, studentName varchar ) CREATE TABLE teacher ( teacherID int PRIMARY KEY, teacherName varchar ) CREATE TABLE student_teacher ( studentID int, teacherID int, PRIMARY KEY(studentID, teacherID), FOREIGN KEY(studentID) REFERENCES student(studentID), FOREIGN KEY(teacherID) REFERENCES teacher(teacherID) )
SemantEducaTrix, the most recent Semantic Web company to burst into the educational software market, is mapping the school system's relational database to RDF / SPARQL. They'd like to access the relationships modeled with this join table as simple links between students and teachers:
ex:student1 ex:studentName "Lee". ex:student2 ex:studentName "Eric". ex:student2 ex:studentName "Juan".
ex:teacher1 ex:teacherName "Michael". ex:teacher2 ex:teacherName "Ahmed". ex:teacher3 ex:teacherName "Marcelo".
ex:student1 ex:has_teacher ex:teacher1, ex:teacher 2 ; ex:student2 ex:has_teacher ex:teacher3 ; ex:student3 ex:has_teacher ex:teacher2 ; ...
Value-based type specification
The TEACHERS table, see above, has a Classification column:
+------------+-----------------+--- ... ---+ | TeacherId | Classification | ... | +------------+-----------------+--- ... ---+ | 1 | History | | | 2 | Physics | | | 3 | Music | | +------------+-----------------+-----------+
SemantEducaTrix wants to instantiate these teachers as different {{{rdf:type}}}s depending on the value in the Classification column:
ex:teacher1 a ex:HistoryTeacher . ex:teacher2 a ex:PhysicsTeacher . ex:teacher3 a ex:MusicTeacher .