


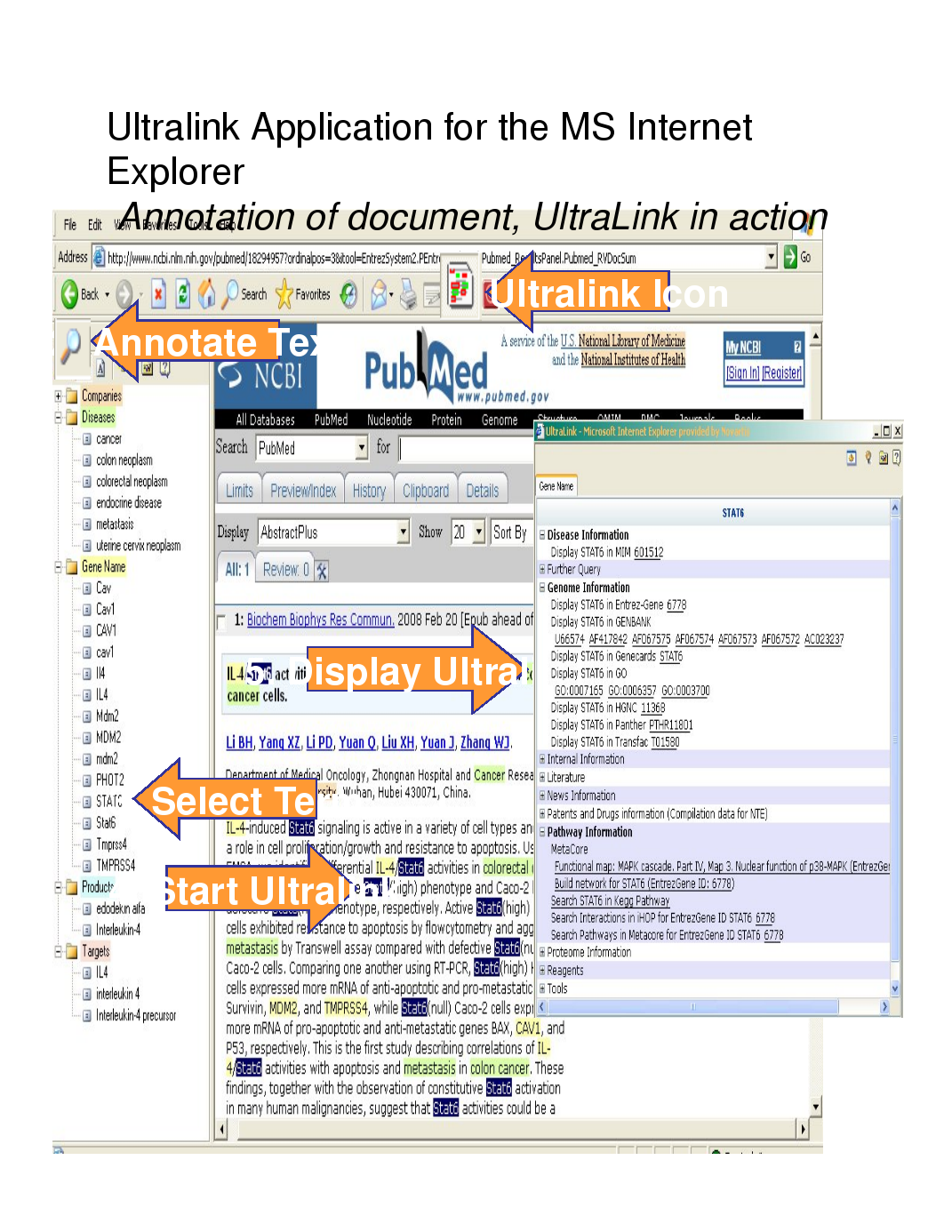
Presentation given at Conference on Semantics in Healthcare & Life Sciences (C-SHALS) 2008, in Boston, USA, on the 5th of March, 2008.
Follow along at http://www.w3.org/2008/Talks/0305-C-SHALS/.
 This work is licensed under a
Creative Commons Attribution 3.0 License,
with attribution to W3C.
This work is licensed under a
Creative Commons Attribution 3.0 License,
with attribution to W3C.
Using the Semantic Web: Precise Answers to Complex Questions:
<?xml version="1.0"?>
<ClinicalDocument transformation="hl7-rim-to-pomr.xslt">
<recordTarget>
<patientRole>
<patientPatient>
<name>
<given>Henry</given>
<family>Levin</family>
</name>
<administrativeGenderCode code="M"/>
<birthTime value="19320924"/>
</patientPatient>
</patientRole>
</recordTarget>
<component>
<StructuredBody>
<Observation>
<code displayName="Cuff blood pressure"/>
<effectiveTime value="200004071430"/>
<targetSiteCode displayName="Left arm"/>
<entryRelationship typeCode="COMP">
<Observation>
<effectiveTime value="200004071530"/>
<value value="132" unit="mm[Hg]"/>
</Observation>
</entryRelationship>
</Observation>
<Observation>
<code displayName="Cuff blood pressure"/>
<effectiveTime value="200004071530"/>
<targetSiteCode displayName="Left arm"/>
<entryRelationship typeCode="COMP">
<Observation>
<code displayName="Systolic BP"/>
<effectiveTime value="200004071530"/>
<value value="135" unit="mm[Hg]"/>
</Observation>
</entryRelationship>
<entryRelationship typeCode="COMP">
<Observation>
<code displayName="Diastolic BP"/>
<effectiveTime value="200004071530"/>
<value value="88" unit="mm[Hg]"/>
</Observation>
</entryRelationship>
</Observation>
</StructuredBody>
</component>
</ClinicalDocument>

<http://thefigtrees.net/lee/id#lee> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://xmlns.com/foaf/0.1/Person> . <http://thefigtrees.net/lee/id#lee> <http://xmlns.com/foaf/0.1/name> "Lee Feigenbaum" . <http://thefigtrees.net/lee/id#lee> <http://xmlns.com/foaf/0.1/homepage> <http://thefigtrees.net/lee/> .
... is more succinctly represented as:
@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> .
@prefix foaf: <http://xmlns.com/foaf/0.1/> .
<http://thefigtrees.net/lee/id#lee> rdf:type foaf:Person ;
foaf:name "Lee Feigenbaum" ;
foaf:homepage <http://thefigtrees.net/lee/> .
_:p1 a galen:Patient ;
foaf:family_name "Levin" ;
foaf:firstName "Henry" .
_:c1a edns:patient _:p1 ;
edns:screeningBP [
a cpr:clinical-examination ;
dc:date "2000-04-07T15:30:00" ;
edns:systolic [
a galen:AbsoluteMeasurement ;
ex:unit "mm[Hg]" ;
r:value "132" ;
skos:prefLabel "Systolic BP"
] ;
edns:diastolic [
a galen:AbsoluteMeasurement ;
ex:unit "mm[Hg]" ;
r:value "86" ;
skos:prefLabel "Diastolic BP"
] ;
edns:location snomed:_66480008 ; # SNOMED:left arm
edns:posture snomed:_163035008 # SNOMED:sitting
] .
| There is a blood-pressure examination of a patient named Henry Levin. The examination was on 7-April-2000 at 3:30pm and was conducted on the patient's left arm while he was sitting. The examination resulted in a systolic blood pressure measurement of 132 and a diastolic measurement of 86. |
SPARQL is the query language of the Semantic Web. It lets us:
| ?artist | ?album | ?times_platinum |
|---|---|---|
| Michael Jackson | Thriller | 27 |
| Led Zeppelin | Led Zeppelin IV | 22 |
| Pink Floyd | The Wall | 22 |
A triple pattern is an RDF triple that can have variables in any of the subject, predicate, or object positions.
Examples:
We can combine more than one triple pattern to retrieve multiple values and easily traverse an RDF graph:
SPARQL lets us query different RDF graphs in a single query. Consider movie reviews:
GRAPH <http://example.org/reviews/rogerebert> {
ex:atonement rev:hasReview ?review .
?review rev:rating ?rating .
}
GRAPH <http://example.org/reviews/rogerebert> {
?movie rev:hasReview ?rev1 .
?rev1 rev:rating ?ebert .
}
GRAPH <http://example.org/reviews/me> {
?movie rev:hasReview ?rev2 .
?rev2 rev:rating ?me .
}
GRAPH ?reviewer_graph {
?review rev:rating 10 .
}
Besides selecting tables of values, SPARQL allows three other types of queries:
SELECT and ASK results can be returned as XML or JSON. CONSTRUCT and DESCRIBE results can be returned via any RDF serialization (e.g. RDF/XML or Turtle).
The SPARQL Protocol is a simple method for asking and answering SPARQL queries over HTTP. A SPARQL URL is built from three parts:
http://example.org/sparql?named-graph-uri=http%3A%2F%2Fexample.orgm%2F reviews%2Febert&query=SELECT+%3Freview_graph+WHERE+%7B%0D%0A++GRAPH+%3Frev iew_graph+%7B%0D%0A+++++%3Freview+rev%3Arating+10+.%0D%0A++%7D%0D%0A%7D
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX edns: <http://www.loa-cnr.it/ontologies/ExtendedDnS.owl#>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX galen: <http://www.co-ode.org/ontologies/galen#>
PREFIX r: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX snomed: <http://termhost.example/SNOMED/>
SELECT ?date ?sys ?dias ?position {
?p r:type galen:Patient ;
foaf:family_name "Levin" ;
foaf:firstName "Henry" .
?c edns:patient ?p ;
edns:screeningBP ?scr .
?scr dc:date ?date ;
edns:systolic [ r:value ?sys ] ;
edns:diastolic [ r:value ?dias ] ;
edns:posture ?position .
} ORDER by ?date
The sample query can be run against this sample data.
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX edns: <http://www.loa-cnr.it/ontologies/ExtendedDnS.owl#>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX galen: <http://www.co-ode.org/ontologies/galen#>
PREFIX r: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX snomed: <http://termhost.example/SNOMED/>
SELECT ?date ?sys ?dias {
?p r:type galen:Patient ;
foaf:family_name "Levin" ;
foaf:firstName "Henry" .
?c edns:patient ?p ;
edns:screeningBP ?scr .
?scr dc:date ?date ;
edns:systolic [ r:value ?sys ] ;
edns:diastolic [ r:value ?dias ] ;
edns:posture snomed:_163035008 . # SNOMED:sitting
} ORDER by ?date
The sample query can be run against this sample data.
GRDDL (Gleaning Resource Descriptions from Dialects of Languages) is a way to boostrap RDF out of XML and in particular XHTML data by explicitly indicating transformations from RDF to XML. GRDDL relies on:
<?xml version="1.0"?>
<ClinicalDocument transformation="hl7-rim-to-pomr.xslt">
<recordTarget>
<patientRole>
<patientPatient>
<name>
<given>Henry</given>
<family>Levin</family>
</name>
<administrativeGenderCode code="M"/>
<birthTime value="19320924"/>
</patientPatient>
</patientRole>
</recordTarget>
<component>
<StructuredBody>
<Observation>
<code displayName="Cuff blood pressure"/>
<effectiveTime value="200004071430"/>
<targetSiteCode displayName="Left arm"/>
<entryRelationship typeCode="COMP">
<Observation>
<effectiveTime value="200004071530"/>
<value value="132" unit="mm[Hg]"/>
</Observation>
</entryRelationship>
</Observation>
<Observation>
<code displayName="Cuff blood pressure"/>
<effectiveTime value="200004071530"/>
<targetSiteCode displayName="Left arm"/>
<entryRelationship typeCode="COMP">
<Observation>
<code displayName="Systolic BP"/>
<effectiveTime value="200004071530"/>
<value value="135" unit="mm[Hg]"/>
</Observation>
</entryRelationship>
<entryRelationship typeCode="COMP">
<Observation>
<code displayName="Diastolic BP"/>
<effectiveTime value="200004071530"/>
<value value="88" unit="mm[Hg]"/>
</Observation>
</entryRelationship>
</Observation>
</StructuredBody>
</component>
</ClinicalDocument>
<xsl:template match="rim:ClinicalDocument[rim:recordTarget/ rim:patientRole/rim:patientPatient]"> <cpr:patient-record> <xsl:apply-templates select="rim:effectiveTime"/> <xsl:apply-templates select="rim:recordTarget/ rim:patientRole/rim:patientPatient"/> <xsl:for-each select="rim:author/ rim:assignedAuthor/rim:assignedPerson"> <foaf:maker> <foaf:Person> <xsl:apply-templates select="rim:name"/> </foaf:Person> </foaf:maker> </xsl:for-each> <xsl:apply-templates select="rim:component"/> </cpr:patient-record> </xsl:template> <xsl:template match="rim:name/rim:family"> <foaf:family_name><xsl:value-of select="."/></foaf:family_name> </xsl:template> <xsl:template match="rim:name/rim:given"> <foaf:firstName><xsl:value-of select="."/></foaf:firstName> </xsl:template> <xsl:template match="rim:patientPatient"> <edns:about> <galen:Patient> <xsl:apply-templates select="rim:name"/> </galen:Patient> </edns:about> </xsl:template>
<xsl:template match="rim:ClinicalDocument[rim:recordTarget/ rim:patientRole/rim:patientPatient]"> <cpr:patient-record> <xsl:apply-templates select="rim:effectiveTime"/> <xsl:apply-templates select="rim:recordTarget/ rim:patientRole/rim:patientPatient"/> <xsl:for-each select="rim:author/ rim:assignedAuthor/rim:assignedPerson"> <foaf:maker> <foaf:Person> <xsl:apply-templates select="rim:name"/> </foaf:Person> </foaf:maker> </xsl:for-each> <xsl:apply-templates select="rim:component"/> </cpr:patient-record> </xsl:template>
<xsl:template match="rim:name/rim:family"> <foaf:family_name><xsl:value-of select="."/></foaf:family_name> </xsl:template> <xsl:template match="rim:name/rim:given"> <foaf:firstName><xsl:value-of select="."/></foaf:firstName> </xsl:template> <xsl:template match="rim:patientPatient"> <edns:about> <galen:Patient> <xsl:apply-templates select="rim:name"/> </galen:Patient> </edns:about> </xsl:template>
<xsl:template match="rim:ClinicalDocument[rim:recordTarget/ rim:patientRole/rim:patientPatient]"> <cpr:patient-record> <xsl:apply-templates select="rim:effectiveTime"/> <xsl:apply-templates select="rim:recordTarget/ rim:patientRole/rim:patientPatient"/> <xsl:for-each select="rim:author/ rim:assignedAuthor/rim:assignedPerson"> <foaf:maker> <foaf:Person> <xsl:apply-templates select="rim:name"/> </foaf:Person> </foaf:maker> </xsl:for-each> <xsl:apply-templates select="rim:component"/> </cpr:patient-record> </xsl:template>
<xsl:template match="rim:name/rim:family"> <foaf:family_name><xsl:value-of select="."/></foaf:family_name> </xsl:template> <xsl:template match="rim:name/rim:given"> <foaf:firstName><xsl:value-of select="."/></foaf:firstName> </xsl:template>
<xsl:template match="rim:patientPatient"> <edns:about> <galen:Patient> <xsl:apply-templates select="rim:name"/> </galen:Patient> </edns:about> </xsl:template>
<xsl:template match="rim:ClinicalDocument[rim:recordTarget/ rim:patientRole/rim:patientPatient]"> <cpr:patient-record> <xsl:apply-templates select="rim:effectiveTime"/> <xsl:apply-templates select="rim:recordTarget/ rim:patientRole/rim:patientPatient"/> <xsl:for-each select="rim:author/ rim:assignedAuthor/rim:assignedPerson"> <foaf:maker> <foaf:Person> <xsl:apply-templates select="rim:name"/> </foaf:Person> </foaf:maker> </xsl:for-each> <xsl:apply-templates select="rim:component"/> </cpr:patient-record> </xsl:template> <xsl:template match="rim:name/rim:family"> <foaf:family_name><xsl:value-of select="."/></foaf:family_name> </xsl:template> <xsl:template match="rim:name/rim:given"> <foaf:firstName><xsl:value-of select="."/></foaf:firstName> </xsl:template>
<xsl:template match="rim:patientPatient"> <edns:about> <galen:Patient> <xsl:apply-templates select="rim:name"/> </galen:Patient> </edns:about> </xsl:template>
XSLT courtesy of Chimezie Ogbuji
GRDDL can extract RDF from both XML and (X)HTML.
| Patient | Systolic BP | Diastolic BP |
|---|---|---|
| Henry Levin | 132 | 86 |
| ... | ... | ... |
<html> <head profile="http://www.w3.org/2003/g/data-view"> <title>Clinical Study 8B1a: Patient BP</title> <link rel="transformation" href="bp-html-to-pomr.xslt" /> </head> ...
Content publisher provides XML or XHTML document that does one of:
Content consumers make use of GRDDL by doing the following:
Some SPARQL engines can directly query GRDDL source documents.
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX edns: <http://www.loa-cnr.it/ontologies/ExtendedDnS.owl#>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX galen: <http://www.co-ode.org/ontologies/galen#>
PREFIX r: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT ?date ?sys ?dias ?location ?position {
?p r:type galen:Patient ;
foaf:family_name "Levin" ;
foaf:firstName "Henry" .
?c edns:patient ?p ;
edns:screeningBP ?scr .
?scr dc:date ?date ;
edns:systolic [ r:value ?sys ] ;
edns:diastolic [ r:value ?dias ] ;
edns:location ?location ;
edns:posture ?position .
}
The actual query against the actual XML source is more complex.
What other musicians are based in Seattle?
Find me all movies that run longer than 5 hours.
| attribute | specifies | attribute | specifies | |
|---|---|---|---|---|
| @about | subjects | @property | predicate relating subject to literal content | |
| @href | objects, clickable | @rel | predicate relating subject to resources (@href, @src) | |
| @src | objects, embedded | @rev | predicate relating resources to subject in reverse | |
| @resource | objects, not clickable | @content | Object of triple (instead of element content) | |
| @instanceof | RDF types | @datatype | literal values' data types |
For more, see the RDFa primer or the RDFa specification.
InChI is a textual identifier for chemical substances. Consider inchi.html:
<table> <tr> <th>Familiar name</th><th>InChI</th> </tr><tr> <td>Methane</td> <td about="http://example.org/methane" property="chem:inchi" xmlns:chem="http://www.blueobelisk.org/chemistryblogs/"> InChI=1/CH4/h1H4 </td> ...
This RDFa encodes the single RDF triple:
<http://example.org/methane> chem:inchi "InChI=1/CH4/h1H4" .
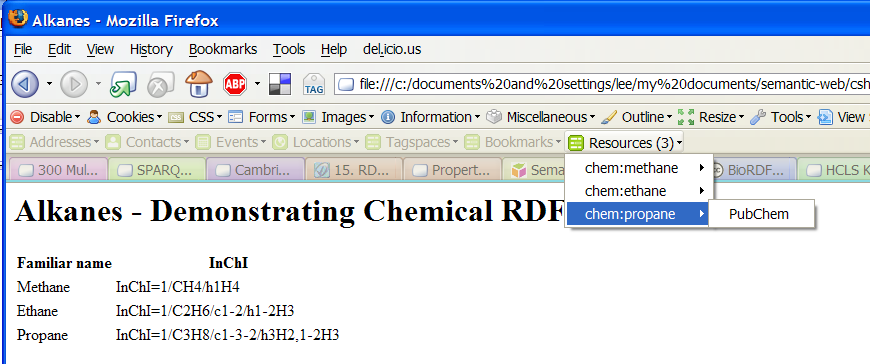
See inchi.html.
There are various ways to query Web pages marked up with RDFa:
# Find propane's InChI string
PREFIX chem: <http://www.blueobelisk.org/chemistryblogs/>
PREFIX ex: <http://example.org/>
SELECT ?inchi
FROM <http://www.w3.org/2007/08/pyRdfa/extract?uri=http://www.w3.org/2008/Talks/0305-C-SHALS/inchi.html>
WHERE {
ex:propane chem:inchi ?inchi .
}
Stages of modeling (frequently in this order):
Class(a:MGHcharlstStPatient partial a:MGHpatient)
Class(a:MGHcharlstStPatient complete
restriction(a:physician
allValuesFrom(unionOf(a:MGHcharlesStOncologist a:MGHcharlesStOptician))))_:MCSO rdfs:subClassOf :MGHpatient . _:MCSO rdfs:subClassOf _:physType . _:physType owl:onProperty :physician . _:physType owl:allValuesFrom _:list1 . _:list1 rdf:first :MGHcharlesStOncologist . _:list1 rdf:rest _:list2 . _:list2 rdf:first :MGHcharlesStOptician . _:list2 rdf:rest rdf:nil . _:MCSO owl:equivalentClass :MGHcharlstStPatient .
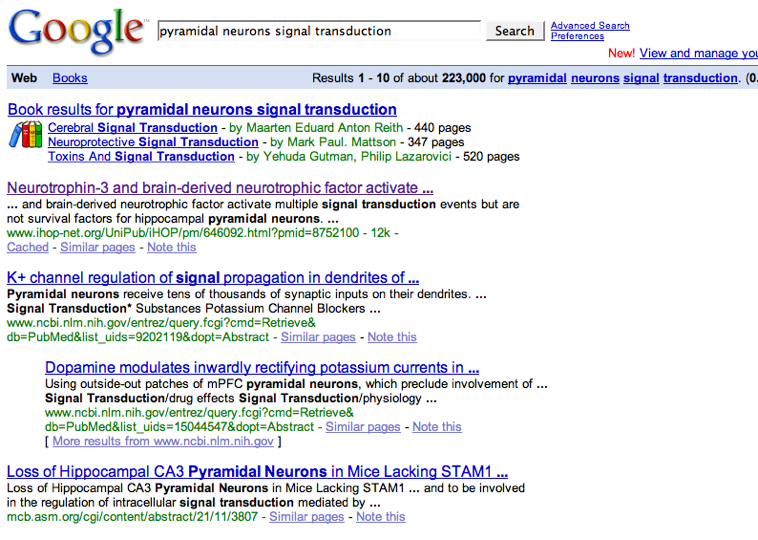
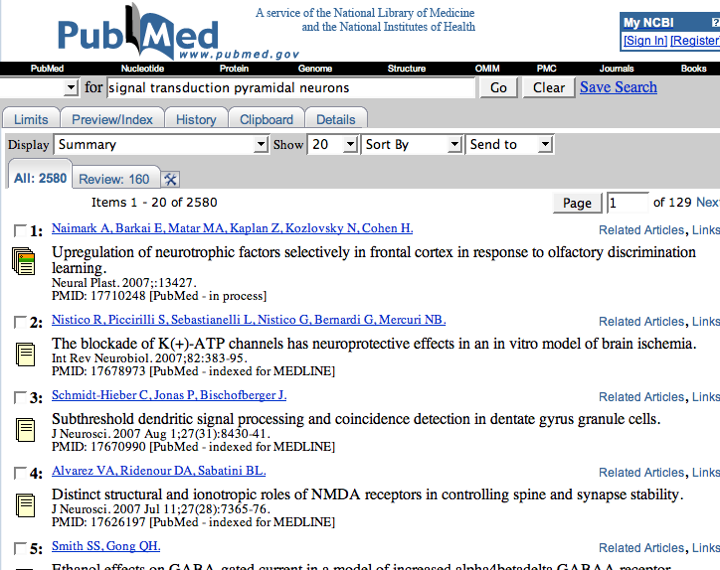
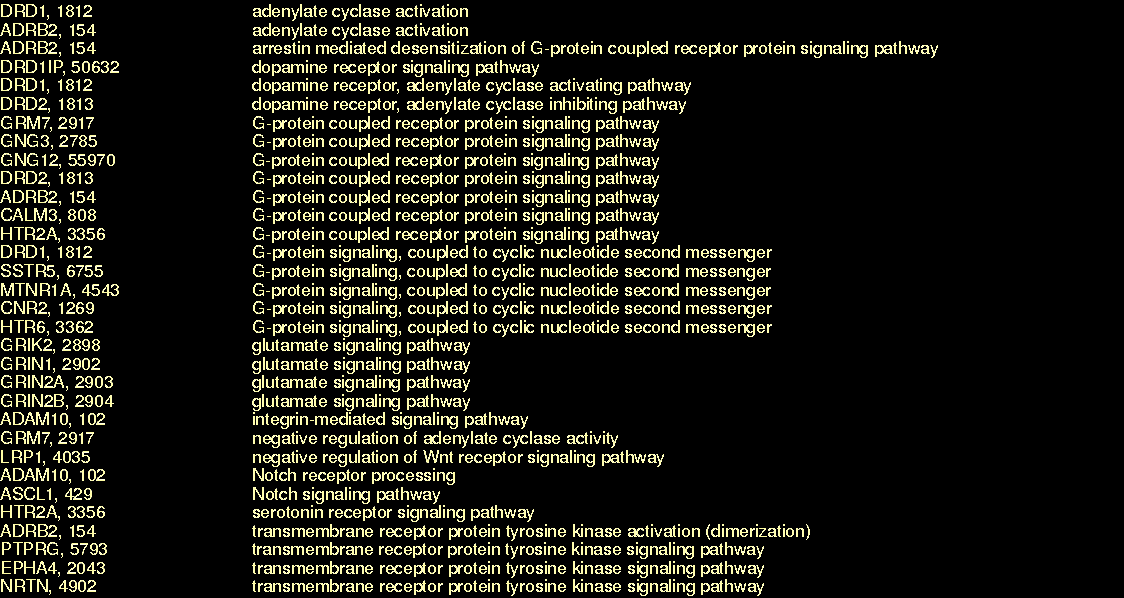
The application of a commercial text mining tool to neuroscience-related PubMed abstracts results in a set of annotations that link MeSH terms to genes (for more details on MeSH, see the table in Data Sources. An article with PubMed id 10698743 mentions ncbi_gene:1812 and that the corresponding PubMed record has a MeSH term mesh:D017966. The following three triples express this:
| pubmedRec:10698743 | sc:has-as-minor-mesh | mesh:D017966 |
| article:10698743 | sc:identified_by_pmid | pubmedRec:10698743 |
| ncbi_gene:1812 | sc:describes_gene_or_gene_product_mentioned_by | article:10698743 |
A set of genes or gene products in human bodies are described by ncbi_gene:1812. Here, we call this set _:equiv1812.
| _:equiv1812 | owl:onProperty | dnaGeneProduct:described_by |
| _:equiv1812 | owl:hasValue | ncbi_gene:1812 |
bySequence:ncbi_gene.1812 is identical to the class _:equiv1812, meaning, it has the same extension (members) but not the same intention (meaning). We assert this identical set because it allows the definition of the gene class to be completely defined by the above two statements (see OWL Web Ontology Language Semantics and Abstract Syntax Section 4. Mapping to RDF Graphs).
| bySequence:ncbi_gene.1812 | owl:equivalentClass | _:equiv1812 |
Using our other supplied constant, we note that adenylate cyclase activation, go:GO_0007190, is part of signal transduction, go:GO_0007166. Note: this simplified query matches only processes that are a sub-process of go:GO_0007166; the actual query, described in §9 Named Graphs, looks also for subclasses. The part_of relationships were inferred from the OWL class restrictions expressed within the shaky line. These are described in §6.1 Modeling Details. The class of functions that are realized_as adenylate cyclase activation is here labeled _:activateAdenylCyclase.
| go:GO_0007190 | obo:part_of | go:GO_0007166 | . |
| _:activateAdenylCyclase | owl:onProperty | ro:realized_as | . |
| _:activateAdenylCyclase | owl:someValuesFrom | go:GO_0007190 | . |
There are many possible classes of substance participating in molecular signaling, one of which (called here _:molecularSignalers_1) is defined by the ability to activate adenyl cyclase.
| _:signalingParticipants_1 | owl:onProperty | ro:has_function | . |
| _:signalingParticipants_1 | owl:someValuesFrom | _:activateAdenylCyclase | . |
The class of proteins in the intersection of _:signalingParticipants_1 and bySequence:ncbi_gene.1812 is here abbreviated protein:p1812_7190_1, though the actual identifier is protein:product_of_ncbi_gene.1812_that_participates_in_GO_0007190_fbc49f20524727a24c7b7effa29bad4a. Note: the Venn diagram reveals that this set is potentially empty (like the intersection of cars and ice cream stands), theoretically permitting the query to range over pairs of gene/process that aren't related through any known protein. However, OWL-DL reasoners will not infer new classes, so the proteins in the intersection of ncbi_gete:1812 and the substances participating in molecular signalling is restricted to the set which have already been entered into the knowledgebase, e.g. like p1812_7190_1.
| protein:p1812_7190_1 | rdfs:subClassOf | _:signalingParticipants_1 | . |
| protein:p1812_7190_1 | rdfs:subClassOf | bySequence:ncbi_gene.1812 | . |
ncbi_gene:1812 and go:GO_0007190 have human-readable labels.
| ncbi_gene:1812 | rdfs:label | "Entrez Gene record for human DRD1, 1812" |
| go:GO_0007190 | rdfs:label | "adenylate cyclase activation" |
try it (or try the tiny URL for the query)
prefix go: <http://purl.org/obo/owl/GO#> prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> prefix owl: <http://www.w3.org/2002/07/owl#> prefix mesh: <http://purl.org/commons/record/mesh/> prefix sc: <http://purl.org/science/owl/sciencecommons/> prefix ro: <http://www.obofoundry.org/ro/ro.owl#> prefix senselab: <http://purl.org/ycmi/senselab/neuron_ontology.owl#> prefix obo: <http://purl.org/obo/owl/obo#> SELECT ?genename ?processname ?receptor_protein_name WHERE { # PubMeSH includes ?gene_records mentioned in ?articles which are identified by pmid in ?pubmed_records . GRAPH <http://purl.org/commons/hcls/pubmesh> { ?pubmed_record sc:has-as-minor-mesh mesh:D017966 . ?article sc:identified_by_pmid ?pubmed_record . ?gene_record sc:describes_gene_or_gene_product_mentioned_by ?article } # The Gene Ontology asserts that foreach ?protein, ?protein ro:has_function [ ro:realized_as ?process ]. GRAPH <http://purl.org/commons/hcls/goa> { ?protein rdfs:subClassOf ?restriction1 . ?restriction1 owl:onProperty ro:has_function . ?restriction1 owl:someValuesFrom ?restriction2 . ?restriction2 owl:onProperty ro:realized_as . ?restriction2 owl:someValuesFrom ?process . # Also, foreach ?protein, ?protein has a parent class which is linked by some predicate to ?gene_record. ?protein rdfs:subClassOf ?protein_superclass . ?protein_superclass owl:equivalentClass ?restriction3 . ?restriction3 owl:onProperty sc:is_protein_gene_product_of_dna_described_by . ?restriction3 owl:hasValue ?gene_record . # Each ?process (that we are interested in) is a subclass of the signal transduction process. # @@ nested graph constraint GRAPH <http://purl.org/commons/hcls/20070416/classrelations> { { ?process obo:part_of go:GO_0007166 } UNION { ?process rdfs:subClassOf go:GO_0007166 } } } GRAPH <http://purl.org/commons/hcls/gene> { ?gene_record rdfs:label ?genename } GRAPH <http://purl.org/commons/hcls/20070416> { ?process rdfs:label ?processname }}