 |
 |
Contributions from the Semantic Web for Life Sciences Group
BioDASH is a Semantic Web prototype of a Drug Development Dashboard that associates disease, compounds, drug progression stages, molecular biology, and pathway knowledge for a team of users. It is based on the concept of a therapeutic topic model, something that exists in one form or another within the pharmaceutical industry. Since such relevant information usually resides across many intranet database servers and different R&D groups, the challenge is more about leveraging the information one already has in a semantically coherent fashion than about making new data models.
The Semantic Web Initiative defines technologies and methodologies that map directly onto many of the challenges in the life sciences. One pressing need is collecting and representing complex forms of information in an intelligent, flexible form so that it is interpretable by software as well as viewable by humans. Another important need is to be able to gain important insights or make critical decisions based on an aggregation of information that may share common entities, such as molecules, diseases, and intellectual property.
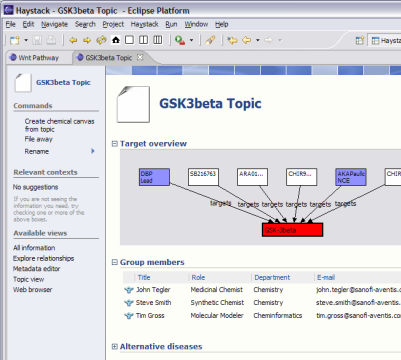
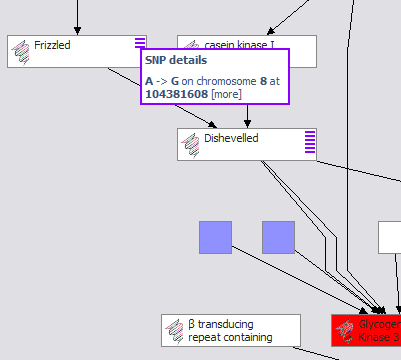
The demonstration focuses on an investigation into the therapeutic value of GSK3beta (glycogen synthase kinase 3 beta), a regulatory enzyme associated with multiple diseases, including diabetes type 2 as well as Alzheimer’s. Several screened compounds are known antagonists of GSK3beta and are available. GSK3beta participates in a variety of important pathways (WNT, Insulin Receptor Singalling), but little is known about the exact roles these play in different tissues, or how they work together. [more on GSK3 beta…]
The BioDASH demonstration is built on Haystack, which is an extensible Semantic Web Browser developed by the Haystack research group at the MIT Computer Science and Artificial Intelligence Laboratory. Haystack is a plug-in for the Eclipse platform and runs on Windows, Linux, and Mac OS X. More information on the use of Semantic Web Browsers in life sciences applications can be found here.
Click here for instructions to download the demonstration onto your own machine. Alternatively, click here to walk through the demonstration.
In the BioDASH demo, information is provided about drug targets and compounds (public sources), which are linked together in the context of a disease topic and a team (see GSK3beta.rdf). A data model is presented that maps Compounds to their drug development role (ChemicalEntities):

All chemical properties are attached to the Compound, while assays and the current drug development stage are associated with the ChemicalEntity. This neatly separates the universal physico-chemical characteristics from those that are business-context specific.
A target is defined similarly as the role for a gene or protein in the context of drug development:

Common biological knowledge is attached to Gene entities, while knowledge specific to target identification, validation, therapeutic mechanisms, compound interactions, and toxic side-effects are associated with a Target. Both Compounds and Chemical Entities can “target” a Gene Target (see above figure). This allows the aggregation and viewing of putative drug relations between a target and any set of compounds, merged from in-house drug development projects, academic research, or competitors.
The basic model provided here offers enough of an initial scaffold to connect information of compounds and chemical libraries with biological entities such as genes, proteins, and pathways, in the context of a drug development project associated with one or more therapeutic areas (primary and alternative). As a real-world project progresses, its information, as well as hypotheses for this topic, can expand or be amended to this topic model. Since the data for this topic view can reside in multiple locations in different data models, the system described can be configured to work with any basic set of drug discovery data systems, aggregating select data through the intranet to offer a dashboard view of drug development with scalable degrees of granularity.
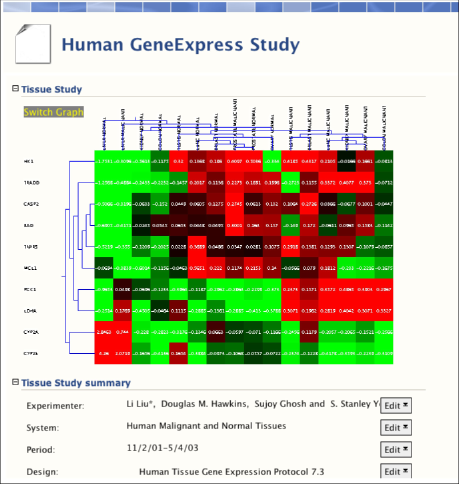
BioDASH is currently being expanded to handle gene expression data, merged with clinical, histological, and toxicity findings, all linked through RDF-OWL:
Example data element in N3:
${ rdf:type ls:GE_Cell;
ls:probeHub gl:CASP2;
ls:GE_Log_Ratio "-0.0677";
ls:conditionHub gl:KIDNEY_MALIGNANT;
ls:histology <urn:lsid:hpr.se:hpr:CASP2_KIDNEY>
} ;
Gene expression data from malignant and normal tissues based on GeneLogic's GeneExpress database were obtained from the work of Liu et al., 2003. The expression table files were converted into RDF using an Excel script.
Aggregates of biological and molecular data will be used to create new knowledge repositories for use in medical research and drug discovery.