HCLSIG/CDS
Note: This task force has been merged into the Clinical Pharmacogenomics task force.
W3C Task Force: Clinical Decision Support for Personalized Medicine
Lead: Matthias Samwald
Teleconference
This task force has telephone conferences the 2nd and 4th Thursdays of each month, at 11am EST (17:00 Central European Time, 8am Pacific Time Zone). Feel free to join these calls!
Elevator pitch
On average, it takes over a decade and a billion Euro to develop a new drug. That is because most drugs that look promising at first turn out not to work for many of the patients, and, even worse, some of the patients are hurt by certain drugs. When a drug is finally on the market, it might take another decade for it to be known widely enough to be correctly prescribed to those patients who actually need it. That is a tragedy for lots of patients, and a lot of money lost. We are using novel information technologies to build a knowledge base from information found on the web, including data about drugs, patients, genetics, diagnostic tests and other things. We hope that when that knowledge base gets integrated into current IT systems, medical doctors and drug developers will be better able to target drug treatments to those patients who actually benefit from it.
Long version
Medical practitioners face unmet information needs for many cases they encounter in daily practice. The predicted shift towards genomic, personalized medicine comes with the promise of making it possible to choose the best available treatment for each patient, but it will also make the clinical decision process even more complex. To bring these new diagnostic and therapeutic developments to their full potential, computer-based clinical decision support systems need to become an integral part of the clinical environment. The World Wide Web and biomedical databases contain rich information that could be used to create such clinical decision support systems. However, the information in these sources is fragmented, of varying quality and often too complex to be used in the busy routine of clinical practice.
The main goal of the task force on Medical Decision Support is to use the Semantic Web technology stack of the W3C to facilitate the creation of an integrated, openly available and trustworthy infrastructure of biomedical data and services that can be used to provide clinical decision support for genomic, personalized medicine.
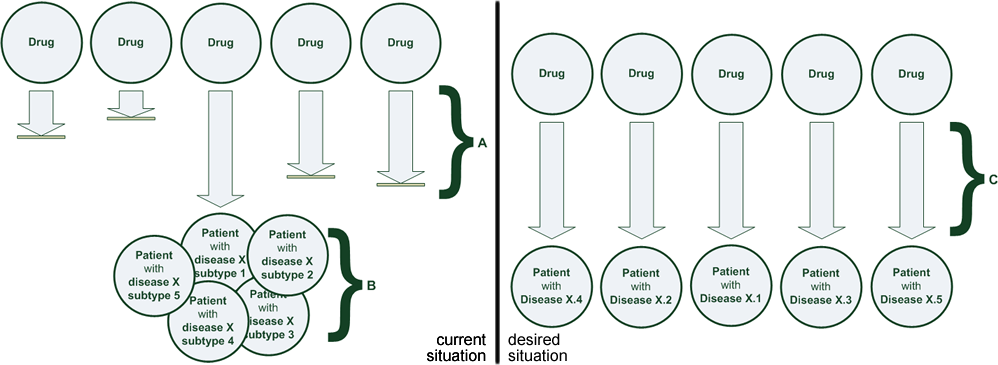
(A) Many promising therapies are found ineffective or unsafe during clinical trials or clinical practice. The establishment of a novel therapy now takes > 17 years and costs > 1 billion € until it reaches patients. (B) The variability in effectiveness and safety among patient subtypes is very large. Approved therapies are only effective for a fraction of the patient population. (C) Adoption of sophisticated clinical decision support tools for personalizing treatment can help medical professionals find the right drug at the right dose for each patient type, and pharmaceutical developers to bring drugs to market that target specific patient types.
Current use cases
The work on each use-case project is intended to be very goal-centric and last up to twelve months, but not longer. New use cases will be proposed on-the-fly. Components developed in each use-case project will be integrated into a coherent system of interoperable ontologies and software (see integration plan below). This process will ensure that the development will be agile and open to new participants and challenges.
1) OWL and SPARQL Rules for Pharmacogenomic Clinical Decision Support
Convert pharmacogenomic and pharmacological datasets to RDF/OWL and map them to the extended Translational Medicine Ontology (developed in cooperation with the Pharmacogenomics task force) as well as the Linked Open Drug Data resources. Explore how SPARQL rules / SPARQL inferencing notation (SPIN) can be used to generate alerts and recommendations to optimize pharmaceutical treatment of individual patients. Explore how the RDF/OWL technology stack can assist the interpretation of genetic patient data for applying pharmacogenomic knowledge in realistic clinical settings.
2) Embedding semantic technologies in clinical environments (alignment with HL7 and OpenCDS)
Explore how RDF/OWL datasets and tools can be used in realistic medical settings and integrated into existing clinical workflows and hospital information systems (e.g., those created by Siemens, Agfa, or VA). As part of this effort, integration with the OpenCDS platform (based on HL7 standards) will be evaluated and collaboration with the OpenCDS community will be established.
3) Biomedical Linked Data for targeted cancer therapy
Combine existing linked data from CancerCommons, molecular pathways in BioPAX format, Drugbank, OMIM and others with newly created linked data to create a Linked Data cloud for targeted cancer therapy, generating an automatically updated 'living review' of current knowledge and data in molecular cancer therapy.
Integration plan
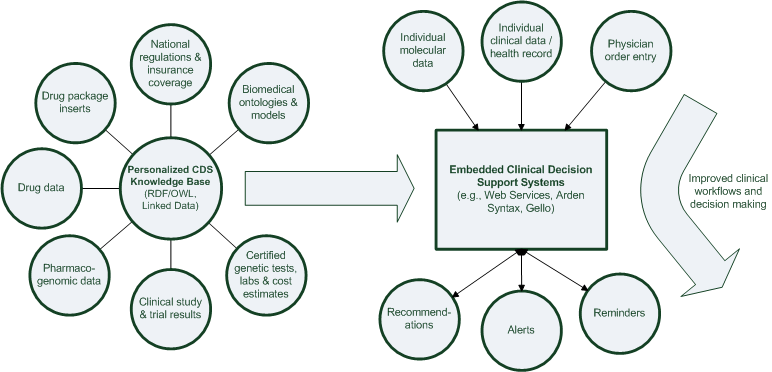
Detail pages
Recommended reading
Trusheim MR, Berndt ER, Douglas FL: Stratified medicine: strategic and economic implications of combining drugs and clinical biomarkers. Nat Rev Drug Discov 2007, 6:287-293.
Eichler H-G, Abadie E, Breckenridge A, Flamion B, Gustafsson LL, Leufkens H, Rowland M, Schneider CK, Bloechl-Daum B: Bridging the efficacy–effectiveness gap: a regulator’s perspective on addressing variability of drug response. Nat Rev Drug Discov 2011, 10:495-506.
Relling MV, Klein TE: CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 2011, 89:464-467.
Kawamoto K: Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ 2005, 330:765-0.
Luciano JS, Andersson B, Batchelor C, Bodenreider O, Clark T, Denney CK, Domarew C, Gambet T, Harland L, Jentzsch A, Kashyap V, Kos P, Kozlovsky J, Lebo T, Marshall SM, McCusker JP, McGuinness DL, Ogbuji C, Pichler E, Powers RL, Prud’hommeaux E, Samwald M, Schriml L, Tonellato PJ, Whetzel PL, Zhao J, Stephens S, Dumontier M: The Translational Medicine Ontology and Knowledge Base: driving personalized medicine by bridging the gap between bench and bedside. J Biomed Sem 2011, 2:S1.
Samwald M, Jentzsch A, Bouton C, Kallesoe CS, Willighagen E, Hajagos J, Marshall MS, Prud’hommeaux E, Hassanzadeh O, Pichler E, Stephens S: Linked open drug data for pharmaceutical research and development. J Cheminf 2011, 3:19.
Samwald M, Stenzhorn H, Dumontier M, Marshall MS, Luciano J, Adlassnig K-P: Towards an interoperable information infrastructure providing decision support for genomic medicine. In Proceedings of Medical Informatics Europe 2011 (MIE2011). 2011.
Links
INBIOMEDvision coordination and support action
STRIDE (Stanford Translational Research Integrated Database Environment)