The pharmaceutical industry is facing increasing pressures to deliver better medicines, faster. There will be many approaches needed to fulfil this aim, and one of these concerns the re-use of existing marketed drugs for new indications. A particular advantage in this strategy is that both the initial research and safety assessment portions of the drug discovery operation have already been completed, thus saving a large amount of both time and cost.
However, it is not just launched, marketed drugs to which this strategy may be applied. Each year, large pharmaceutical companies can work on 100s of simultaneous projects, each with chemical compounds at different phases in the drug discovery pipeline. Some of these compounds may have progressed through to launched drugs, but many will have not and there are a number of different reasons for this. For instance, the compound may have proved toxic in either laboratory experiments or clinical trials, and as such is no longer pursued. However, a significant number of these compounds may be “safe” but will not have been progressed for other reasons, such as the company decided to stop work in a given disease area, or that the compound didn’t have an appreciable effect on the disease for which it was originally intended. Over time, a formidable set of internal compounds, each with a significant amount of scientific research behind it is built up. Looking at the latest medical research to identify new uses for these compounds is one important way to maximize efficiency and use of previous work within the company.
Although it may seem obvious that ensuring any good quality compound is investigated for any utility, there is a significant problem. Namely, in such a large organization, how do scientists collaborate and identify when an internal compound (which may have been created many years ago) becomes interesting to them—particularly when there are so many different databases and tools which could assist this decision?
Within Pfizer we are using Semantic Web technologies to assist us in this process. Our approach was to keep the primary sources of data in standard relational or XML-database format, but export key “facts” as triples in RDF. In our study, the ability to maintain data in a relational format but combine it in an RDF format gave us the best balance between ease of maintenance and ease of use of the data. As an example of how the RDF-version of the data can be used quickly, and effectively, we utilized MIT’s SIMILE’s Exhibit technology to combine different results sets and facilitate decision making (see below). It should be noted that this use case serves merely to illustrate the overall approach to the problem, and that the data is fictitious and randomised.
In the scenario below, a member of the Allergy & Respiratory therapeutic area wishes to search for new opportunities to utilize the progress made by other teams. They open up the Web page shown in Figure 1 which is centred on the internal portfolio across all therapeutic areas, but then cross referenced with public and private information associating the projects with data relevant to Allergy & Respiratory. The data which populates this table is drawn from a variety of different internal and external systems. Key “facts” are exported from these systems into RDF and this data then passed to Exhibit to generate the overview below.
Using the tool above, the Allergy & Respiratory scientist is able to ask the question “for targets being investigated by other Therapeutic areas, which have any information that suggest they may also be useful for me?”. Achieving this requires aggregating data from databases, with data that has been mapped to ontologies, and with results from data mining experiments. The example above contains information from sources such as OMIM GEO which are extensive public repositories of biological information that can be used to ask the question “does this target have any associations with a relevant disease”. Additional data is found in the “Lit_All” and related columns. This data is the result of large scale text-mining experiments which run continually over the biomedical literature searching for associations between targets and disease mechanisms Armed with this information, the scientist is able to see the external and data mined information in the context of events within the organization which aids in judging which opportunities to move forward quickly.
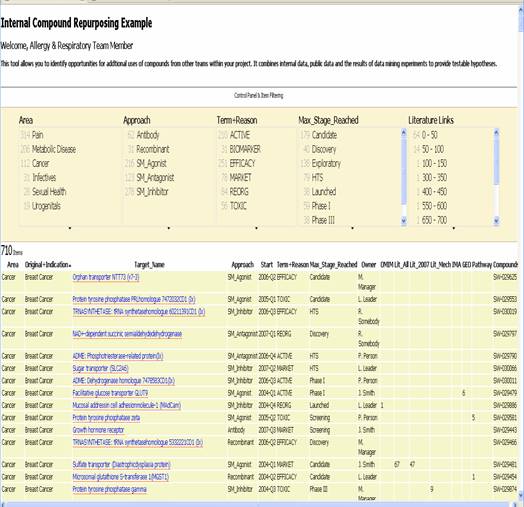
Figure 1: Exhibit-based Internal Collaboration & Repurposing Portal. The portal presents a representation of key data from internal and external sources
In this particular example, the scientist is only interested in projects that are not used for cancer (for safety reasons, perhaps) and so has deselect this in the exhibit filter (see Figure 2). Similarly, they are only interested in small molecule projects, so deselect the biological approaches. They then ask to see only those projects that have terminated for efficacy reasons i.e. in the original project, a non-toxic compound was created, but it simply did not show signs of working for that disease in the models tested. Nevertheless, these compounds form important substrate for repurposing as they may still display promising signs in other diseases.
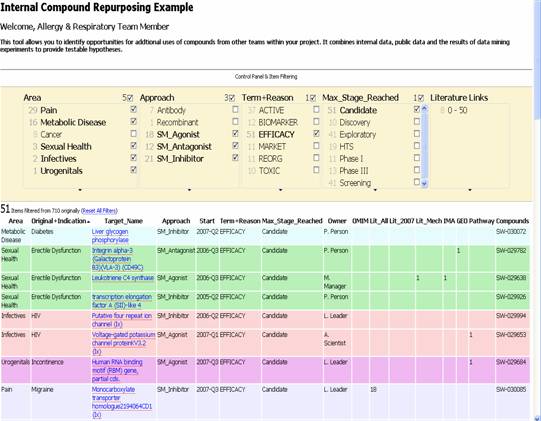
Figure 2: Filtered Opportunity Analysis
From a portfolio of over 700 projects, the scientist has rapidly identified 51 that warrant further investigation and additional filtering could be employed to triage this further. Clicking on the hyperlinked “Target Name” will take the user to the evidence which they can then review and decide on a course of action. These decisions are stored in the system and so can be used as additional columns, removing entries the user has already evaluated from the list. A system based on these techniques is fully in place at Pfizer and is one of the tools used regularly to identify new opportunities.
Our initial work has demonstrated that RDF technology fits well into our data aggregation process. By combining this technology with relational and XML database technology, we can provide powerful systems to enhance drug discovery. It is not necessary to convert every internal and external system to an RDF format. Rather, we look to maintain data in a relational schema (where there are many existing tools) but export key elements into RDF. This allows us to create a “virtual shelf”, from which users can pick and choose different, but compatible datasets for further analysis and visualization. Although technologies such as Exhibit are only just emerging, the ease in which we were able to create a real-life and beneficial application shows that there is great promise for this approach in the future. We were able to build an application platform for delivering this data within just a few hours, and were able to customise these views (for instance, per therapeutic area) very rapidly. This demonstrates a key potential benefit of these technologies, namely that the emphasis (and developer time) is focused on the data, and lightweight applications can be built quickly with minimal overheads. Thus, Semantic web technology provides a means of storing and interpreting any type of data and may be expanded and modified as more data becomes available and if the underlying science changes, allowing us to more rapidly respond to the changing needs of our customers.
© Copyright 2007, Pfizer Ltd.