July 2007

Innovative research institutes rely on the availability of complete and accurate information about new research and development, and it is the business of information providers such as Elsevier to provide the required information in a cost-effective way. It is very likely that the semantic web will make an important contribution to this effort, since it facilitates access to an unprecedented quantity of data. However, with the unremitting growth of scientific information, integrating access to all this information remains a significant problem, not least because of the heterogeneity of the information sources involved - sources which may use different syntactic standards (syntactic heterogeneity), organize information in very different ways (structural heterogeneity) and even use different terminologies to refer to the same information (semantic heterogeneity). The ability to address these different kinds of heterogeneity is the key to integrated access.
Thesauri have already proven to be a core technology to effective information access as they provide controlled vocabularies for indexing information, and thereby help to overcome some of the problems of free-text search by relating and grouping relevant terms in a specific domain. However, currently there is no open architecture which supports the use of these thesauri for querying other data sources. For example, when we move from the centralized and controlled use of EMTREE within EMBASE.com to a distributed setting, it becomes crucial to improve access to the thesaurus by means of a standardized representation using open data standards that allow for semantic qualifications. In general, mental models and keywords for accessing data diverge between subject areas and communities, and so many different ontologies have been developed. An ideal architecture must therefore support the disclosure of distributed and heterogeneous data sources through different ontologies. The aim of the DOPE project (Drug Ontology Project for Elsevier) is to investigate the possibility of providing access to multiple information sources in the area of life science through a single interface.
This approach is sketched in Figure 1 (the letters refer to the figure):
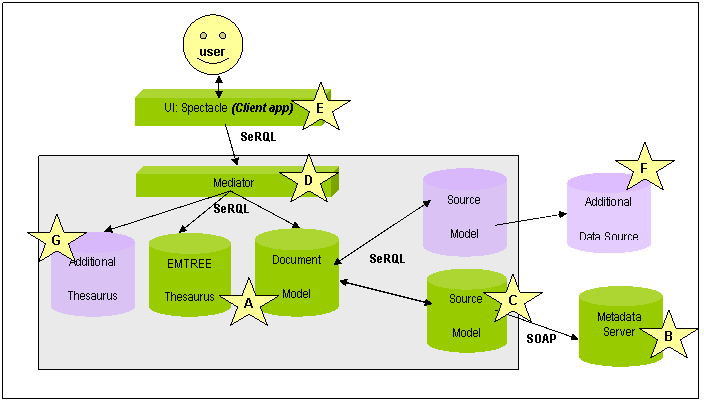
Figure 1: Basic Schematic of the DOPE architecture (protocols and data formats given in bold)
In order to provide the required functionality, a technical infrastructure is needed to mediate between the information sources, thesaurus representation and document metadata stored on the Collexis fingerprint server. Besides the technical integration, the representations of the different information sources have to be integrated on a syntactic and structural level.
In the DOPE prototype, this mediation is implemented using the RDF repository Sesame. On a syntactic level we have achieved interoperability by converting all relevant sources to RDF. In particular, we produced an RDF version of the EMTREE thesaurus. The hierarchy of the thesaurus is represented as an RDF schema class hierarchy enabling us to use the reasoning abilities of Sesame to expand user queries to narrower keywords. The problem of structural heterogeneity between the different sources was addressed using transformations on the RDF representation of information. These transformations have been implemented using the Sesame query language SeRQL, which also supports queries that create an RDF model as output differing structurally from the queried model. These so-called construct-queries are used to communicate with the fingerprint server of the Collexis server. The Collexis Server is a repository of information that is not equipped with RDF-based in- and output facilities. Therefore, an Extractor component is deployed which, through use of the Collexis SOAP interface, converts the available information in an RDF format that is a 1:1 mapping to the original information: the physical model (see Figure 2).
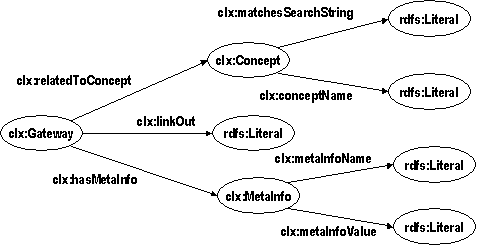
Figure 2: The physical model: an ontology in Collexis terminology
Although the physical model is already in RDF, it is not in the terminology in which the queries are formulated; furthermore, it is less suited to direct merging with different data sources. Therefore, the SeRQL query and transformation language is used to transform the physical model into a logical model. The logical model is based on a subset of the OntoWeb ontology that has been adapted to our purposes. In particular, the representation of author information has been simplified, and the model has been linked to the schema we use to represent the EMTREE thesaurus. This link can be seen in the lower part of Figure 2: each publication is linked to an RDF schema class which represents a preferred term in the thesaurus, and which is further characterized by a label and a relation to similar search strings that is computed on the fly when a query is processed.
A prototype of a user interface client called the “DOPE Browser” has been designed and created. It provides querying and navigation of a collection of documents using thesaurus-based techniques, while hiding much of the complexity of the back-end, such as the existence of multiple data sources, any thesaurus or ontology mapping that may take place, etc. In this system, the user sees a single virtual document collection made navigable using a single thesaurus (EMTREE). Typical document metadata such as e.g. title, authors and journal information is associated with each document. Due to this simplified view on the data, the user interface will be easily reusable on other data sets and thesauri. The DOPE Browser makes use of a thesaurus-driven, interactive visualization technology called the Cluster Map, developed by Aduna, for creating overviews of and getting insight in the available information.
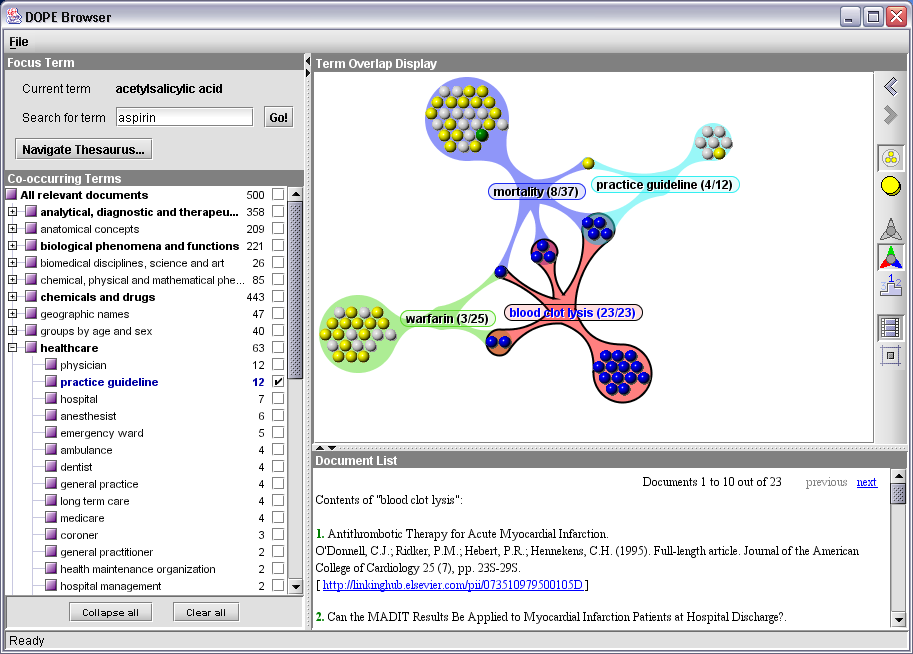
Figure 3: The DOPE Browser
One assumption made during the design is that the EMTREE thesaurus is too large for end users to navigate directly. Researchers typically focus their work on an area that can be described by specific terms nested deep inside a thesaurus. They may have difficulties finding their way to these terms. Apart from the cognitive load, manual navigation of the thesaurus may also be cumbersome simply because of its size. An approach has been followed where the user can quickly focus on a topically related subset of both the document collection and the thesaurus. First, the user selects a single thesaurus keyword. The system then fetches all documents indexed with this target keyword, as well as all the other keywords with which these documents are indexed. These co-occurring keywords are used to provide an interface in which the user can explore the set of documents indexed with the focus keyword.
Suppose a user wants to browse through the existing literature on aspirin. The string “aspirin” can be entered in the text field at the upper left of the figure. The system then consults Sesame for all keywords than are related to this string. It responds with a dialog showing four possible EMTREE terms, asking the user to select one. (This dialog is omitted when there is only one exact match with an EMTREE keyword.) Assuming that the user chooses the keyword “acetylsalicylic acid”, which is the chemical name corresponding with the brand name, this becomes the new focus keyword. The system consults Sesame again and retrieves up to 500 most relevant documents about “acetylsalicylic acid”, including their metadata fields (e.g., titles and authors) and the other keywords with which these documents are indexed. The co-occurring keywords are presented in the tree at the left hand side of the screen, grouped by their facet keyword (the most generic broader keyword, i.e. the root of the tree they belong to). The user can now browse through the tree and check one or more checkboxes that appear alongside the keywords. This action results in a visualisation of their relations and contents at the right hand side of the screen.
Figure 3 shows the state of the interface after the user has checked the terms “mortality”, “practice guideline”, “blood clot lysis” and “warfarin”. The visualization graph shows if and how their document sets overlap. Each sphere in the graph represents an individual document, with its color reflecting the document type, e.g. full article, review article or abstract. The colored edges between keyword and clusters of spheres reveal that those documents are indexed with that keyword. For example, there are 25 documents about warfarin, 22 of them are only labeled with this keyword, two have also been labeled with blood clot lysis, and one is about warfarin, blood clot lysis and mortality. This visualization shows that within the set of documents about aspirin there is some significant overlap between the keywords blood clot lysis and mortality, and that 4 of the practice guidelines documents relate to these two topics as well.
Various ways exist to further explore this graph. The user can click on a keyword or a cluster of articles to highlight their spheres and list the document metadata in the panel at the lower right. Moving the mouse over the spheres reveals the same metadata in a tool tip. The visualizations can also be exported to a clickable image map that can be opened in a web browser.
The use of RDF-based datamodels and exchange syntax has greatly eased the integration of heterogeneous information sources. The EMTREE thesaurus, the Medline database and the Science Direct collection were all developed independently over many years, with separate separate datamodels and syntactic formats. By wrapping the indexed database as an RDF source and by transforming EMTREE into an RDF Schema structure it was possible to integrate these heterogenous sources.
The semantics of the thesaurus is used in the following ways in the functionality of the DOPE system:
Discussions with users about the potential benefits supported the conclusion that the main benefit of the tool is the exploration of a large, mostly unknown information space rather than support for searching for concrete articles. Examples of beneficial applications mentioned by potential end users included: filtering material for preparing lectures about a certain topic, and supporting graduate students in doing literature surveys (e.g. using a “shopping basket” to collect search results). A more advanced potential application that was mentioned was to monitor changes in the focus of the research community. This however would require an extension of the current system with mechanisms for filtering documents based on date of publication as well as advanced visualization strategies for changes that happen over time.
Currently, Aduna and Elsevier are discussing a more widespread adoption of the DOPE prototype in a corporate setting, for use to access various heterogeneous datasets with different, overlapping ontologies. The principal DOPE architecture will be used as a starting point for these investigations. The DOPE prototype which is currently offline will be rebuilt using insights from recent work from both parties.
© Copyright 2007, Elsevier, Aduna, and Vrije Universiteit Amsterdam