To demonstrate the feasibility of applying a Semantic Web aggregation model for a drug discovery problem using basic RDF-OWL tools and methodologies, on both the back-end (servers) and front-end (browsers).We also hope to assess whether specific practices and functionality that are typically challenging to implement through a relational database (RDBM) and/or portal approach lend themselves better to a Semantic Web paradigm.
The GSK3beta Topic items can each be opened and inspected using Haystack—those with external links will allow new data to be aggregated and browsed (e.g., OMIM, Uniprot). These links take advantage of life science identifiers (LSID) specification, resolved into RDF via the lsid.biopathways.org server. The OMIM pages for both Diabetes Type 2 and Alzheimer’s are viewable as HTML within Haystack.
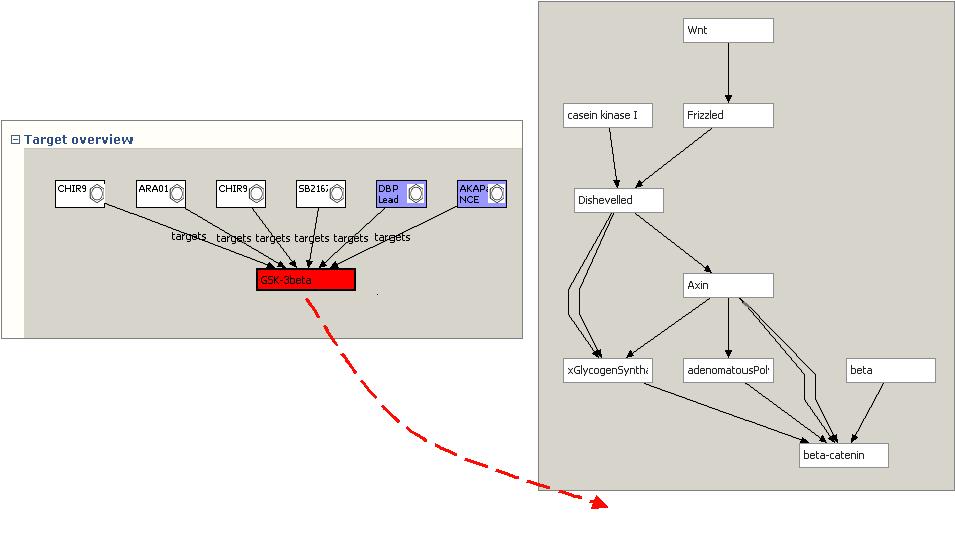
When the WNT pathway view is opened, a BioPAX-RDF file containing the WNT pathway retrieved and rendered to display the signalling cascade. This view is based on a separate semantic model (BioPAXI) from the compound target information. However, the user can choose to drag the target protein (GSK3b) from the Target view onto the Pathway View:

When released, the Pathway View will identify any external references to Uniprot in both the target and pathway graphs. Because all models use Uniprot LSIDs, all that is required is to determine if the same Uniprot LSID URI node is used in multiple cases. If a common LSID node is found, the associated protein node in the pathway is colored red, and the compounds attached to the target are brought across into the pathway view. In addition, if any of the compounds has any other targets that are in the pathway set, that link is also identified using LSIDs, and the edge is rendered. SNP information for each pathway component can also be aggregated from databases such as db-SNP and integrated into the view:
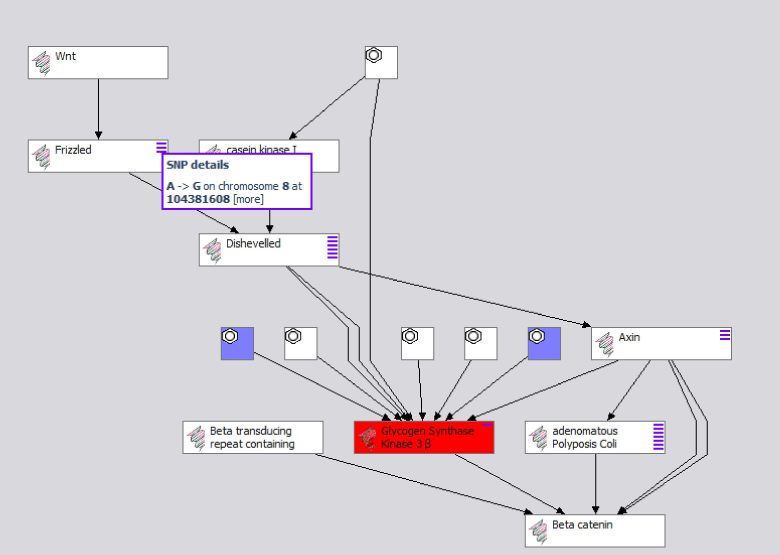
The Pathway view now contains protein and interacting compounds, as well as non-synonymous polymorphisms (purple dashes) linked to individual changes in the protein coding, yet no change to any data models was required! This combined view is based on merged RDF, and therefore could be serialized as such. Each pathway protein, SNP or compound node can be browsed in deeper views as well. If additional information is aggregated to any of these elements (other pathways, additional targets, etc), it will be accessible as well.
Three initial viewing planes or canvases are provided:
This renders the therapeutic topic (in RDF) and aggregates the underlying related information from sources, including Uniprot, OMIM, chemical sources, and project team.
This viewer has lenses that accept BioPAX RDF/XML data and renders just the components in a causal-graph format.
This canvas is used to look at multiple relations between compounds and Chemical Entities. It also manages chemical libraries, and will eventually support
References
Cohen P, Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat Rev Drug Discov. 2004 Jun;3(6):479-87. Review. PMID: 15173837