HCLS/SciPubSPERequirements
Requirements for Experiment Self-publishing Ontology
- **Everyone are welcome to contribute***
Status of this document:
6/18/06 Proposed by AJ Chen
Contributors: (add your name here after you contribute to this document!)
AJ Chen, Bill Bug, Chimezie Ogbuji, Eric Neumann, Matthias Samwald, Sean Martin, Trish Whetzel, Xiaoshu Wang, C. H. Marcondes
General
Publishing information of single experiment as a unit is a simple concept. But, it's also a complicated matter when considering the breath and depth that an experiment may have. For practical reason, it would be helpful to recognize that one ontology may not be appropriate to describe every aspect of a single experiment. Thus, we should expect multiple ontologies will be developed for this purpose of scientific publishing in the future. For now, we will tackle one problem at a time while making sure there is a consistency among the present effort and other future efforts.
An experiment can be described in general terms and/or domain-specific terms. Whether to focus on general description or domain-specific description depends on the applications that the resulted ontology is intended to augment. As described in the task proposal, the focus of the current task is in the area of general description of experiment for the purpose of better data sharing and discovery on the web.
As a good comparison, the task is intended to follow the paths that led to the popular ontologies like FOAF, SKOS and DC. In fact, we should try to use the terms in these existing ontologies as much as possible. Hopefully, the ontology developed by this task will be used as widely as these popular ontologies.
In the following sections, the intended use cases are outlined first. From these use cases, we can then define the requirements for the ontology. Once the requirements are defined, it would be more straight forward to design an ontology in the next step. Everyone are welcome to add additional use cases and requirements directly in this document.
Comment: Bill Bug provideded more fine-grained terms used to describe all the various types of experiments performed by BIRN associated labs. See the discussion thread.
Use Cases
- Any researcher can publish his or her experiment data as single unit of experiment in RDF format using this ontology. Ideally, a new easy-to-use self-publishing tool specifically designed for such purpose will encourage the use of the ontology. However, any ontology editor can be used to create RDF content. Because data in such format can be searched by semantic web search engine, it offers a new channel for data sharing and retrieval that will accelerate scientific discovery as well as increase the researcher's visibility.
- Any tool providers (manufacturers and distributors) can publish product information using this ontology. The products include reagents, materials, instruments, and software that are used by researchers to do their experiments.
- New self-publishing tools can be developed to use the ontology for supporting researchers to self-publish experiment information on the web. Particularly, easy-to-use tools are critical to make the promise of semantic web a reality. Hopefully, an ontology with potentially large user base (all research areas instead of one specific area such as microarray) will be attractive enough to tool developers.
- New search engines can aggregate the experiment information published with the ontology and make the experiment information searchable to everyone on the web.
- Anyone can use the new search engines to find all experiments available on the web. Compared to traditional literature search like Medline, this semantic web search will find much more relevant information with much higher specificity in a much bigger scale. For examples, users can get answers to the following questions: (please add your questions here)
- What experiments have been done on this protein or gene?
- What projects are related to this protein or gene?
- Who are studying this protein or gene? Rank them by the amount of experiments done or by the number of references (i.e. hyperlinks) to their published experiments.
- What hypothesis have people proposed for this protein or gene?
- What are the conclusions people have drawn for this protein or gene?
- What tools/reagents/instrument/protocols have been used in characterize the toxicity of this compound?
- An appropriate Web self-publishing tool can permit author/researchers publishing of scientific articles both as text form human reading and in machine readable format for software agents processing. Knowledge can thus be extracted and represented. Knowledge unit is a proposition corresponding to the hypothesis element of the Scientific Methodology. A hypothesis is a (new) relation between phenomena, expressed as a, for example, a causal relation between concepts in a domain. A hypothesis has the following format that can be extracted and marked up:
<antecedent>to smoke</antecedent>, <type_of_relation>causes</type_of_relation>, <consequent>pulmonary carcinoma</consequent>.
This can permit semantic retrieval queries as:
What other antecedents can cause pulmonary carcinoma? What other diseases to smoke can cause?
Requirements
- The ontology should not be too complicated to use. If a user feels overwhelmed by the large number of parameters required to describe an experiment, this user may hesitate to do it. So, we need to find a good balance: The ontology shoud be as simple as possible as long as it supports the intended applications.
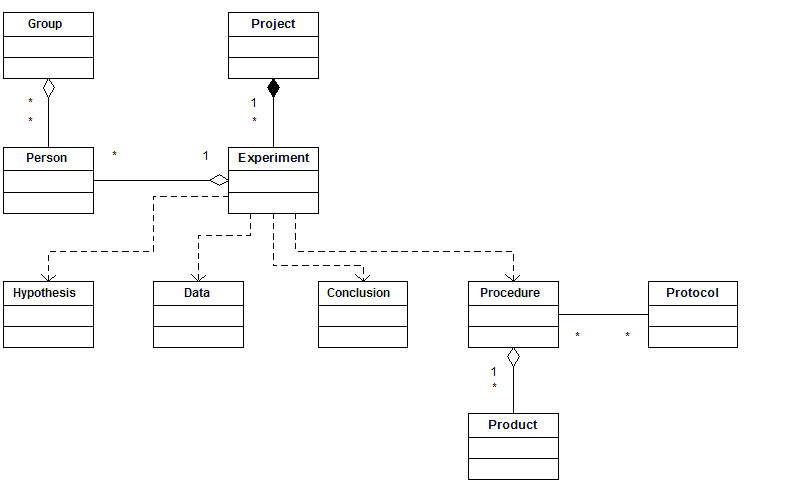
- An experiment can be described by the following objects and properties in order to support the above use cases. Note that it's allowed to use only a subset of these objects and properties to publish any specific experiment.
Properties of Experiment object:
- URI identifier
- Associated project (A Project object)
- Name
- Description
- Hypothesis (Literal or object? Stating the Hypothesis (and model using RDF) that is being tested by the experiment; this includes which citations are supportive or alternative to ones hypothesis. Hypotheses should be defined in terms of authorship (ala DC), what the proposed new concepts is, and what (experimental) fact (or claim) is required to support it. It should also refer to earlier hypotheses either by: a. extension of an earlier tested and supported hypothesis: refinement. b. similarity or congruence with another untested hypothesis: supportive. c. being an alternative to another hypothesis, that will qualify itself through the refutation of the earlier one: refutation. This would allow one to define rules and queries that can traverse the lineage of hypotheses (forwards and backwards, similar to citations), and how one papers work can be related to ongoing work on different fronts that have branched. Reasoning method: theoretical-adductive, experimental-inductive or experimental-deductive)
- Experiment Procedures (A Procedure object)
- protocol used
- Data (Literal or object? possibly as RDF-OWL aggregates and tables.)
- Pointer to data that can't be expressed here.
- Result (Articulating the Results and Conclusions; specifically, whether the experiment refutes or supports the central Hypothesis )
- Discussion (Literal or object?)
- Experiment type or category
- main concepts such as specific proteins or genes, URI as value so that cross-reference becomes possible.
- Special technologies used in the experiment
- Start time
- Finish time
- Location
- Persons who did the experiment (one or more. to indicate different roles of different contributors? +1)
- Related resources
- Related publications
- Homepage
- DataAvailableAt (may be a public repository entry)
- Availability
Properties of Project object:
- URI identifier
- Name
- Description
- Category
- Status
- Owner
- Members (researchers)
- Funding sources
- Homepage
- Related resource
- Related publications
- Start date / Is this necessary ?
- Finish date / " "
Properties of Procedure object:
- URI identifier
- Procedure steps
- Any protocol used in the procedure (A Protocol object)
- Material used
- Equipment used
- Software used
Protocol:
- URI identifier
- Title
- Subject or category
- Procedure steps (A Procedure object)
- References
- Creation time
- Who created it
- Modification time
- From which other protocol has it been derived
- Who modified it
- Owner
- Homepage
Properties of Product object:
- URI identifier
- Name
- Model
- Type
- Specifications
- Manufacturer
- Homepage
Properties of Person object:
- URI identifier
- Full name
- Job Title / Note: Job is not a property of a person, as it may change. It is related to the contribution of a person to a project.
- Salutation
- Working group
- Interests
- Expertise
- Publications
- Weblog
- Homepage
- Contact info
Properties of Group object:
- URI identifier
- Name
- Members
- Organization
- Homepage
- contact info
Properties of Publication object:
- URI identifier
- Role (such as Peer-Reviewed, Electronically-Published, Topic Review, and Follow-up Data; regulatory applications, Common Technical Document)
Additional:
- publishing protocols. which ontology and version?
Note: Hypothesis, Data and Conclusion may be simply represented by string or literal, which is sufficient for search engine application. But, it may offer advantages for some applications if they are represented as object. If anyone has an application that requires object representation for Hypothesis, Data and Conclusion of an experiment, please contribute a use case and define the objects.
Note by MS: It might be useful to re-use the FOAF ontology to represent information about persons and groups (prefarable a version of FOAF that has been adapted to be OWL DL)
Comment by EricN: "Publication" should be a specific concept in SPE, that would serve to be the hub of DC metadata as well as the above experimental data and hypotheses. Different non-disjoint Publication "Roles" could be defined, such as Peer-Reviewed, Electronically-Published, Topic Review, and Follow-up Data. I would also invite the folks interested in Clinical Publications to specify what requirements they feel should be included, (e.g. regulatory applications, Common Technical Document).
Comments by AS: Hypotheses: not all experiments are hypthesis, for istance we may analyze the behaviour of a system to try to reverse engineer it, or just to see what's happening. This does not mean that the experiment has not an object. I think at least an About property is needed in this case, to assert something like: "About metabolism" "About yest" "About shift", I don't know if I give the idea... this are like TAGs or keywords (but from URIs). Netherless in many cases this will be the only available link. And they can be not considered if they don't lead to enough specificity. Or About may link to some proper domain ontology.
Data: not all data can practically (or even must...) fit an RDF file... a pointer to data is mandatory, as well as information on its availability.
I think the associaton of people to projects should be mediate through an object "contribution", o which job title is a property, as well as expertises involved...
Product objects may just be a URI to the vendor+model...
The procedure/protocol link: has a protocol a procedure that has protocol that has procedure... ? This may not be easy to understeand...
Note by C. H. Marcondes: Hypothesis are embedded in a reasoning procedure which implies a reasoning method used in the text of a scientific article. Reasoning methods in articles can be theoretical-adductive, experimental-inductive or experimental-deductive. Theoretical-abductive articles analyze different previous hypothesis, show their faults and limitations and propose a new hypothesis. Experimental-inductive articles propose a hypothesis and develop experiments to test and validate it. Experimental-deductive articles use hypothesis proposed by other researchers and, apply it to a slightly different context and also develop experiments to test and validate it.