HCLS/ClinicalObservationsInteroperability/FDATherapeuticAreaOntologies/LoadingTAOntologiesIntoTBC
Document Conventions
- User actions are in headings - Readers can perform the demo from the table of contents and click on the sections when they need to see screenshots.
- Menu actions separated by "·" - Text like "File·New·Project..." indicates the "File" menu with a submenu of "New" etc.
- Window hints in () - It's easy to lose windows so hints are included in "()". The text "(Window·Show view·Navigator)" shows how to restore the "Navigator pane from Window·Show view submenu.
- back references to earlier actions - Readers can traverse the demo in any order. Repeated actions, like importing a file from the web, have links back to where the action is explained.
TBC demo setup - Creating a project
Top Braid Composer is built on top of Eclipse. Eclipse source files and user interface state are organized in "projects".
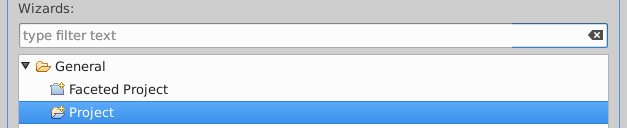
File·New·Project
General·Project, enter "FDA-TA" and select Finish
 This creates a new directory called "FDA-TA" in your Eclipse workspace.
You can look there for the files you import.
If you move files in or out of the project directory, or edit them with an external editor, you may need to "Refresh" the directory (File·Refresh or Right-click on the project in the Navigator pane and select "Refresh").
This creates a new directory called "FDA-TA" in your Eclipse workspace.
You can look there for the files you import.
If you move files in or out of the project directory, or edit them with an external editor, you may need to "Refresh" the directory (File·Refresh or Right-click on the project in the Navigator pane and select "Refresh").
TBC demo 1 - loading all TA Ontologies
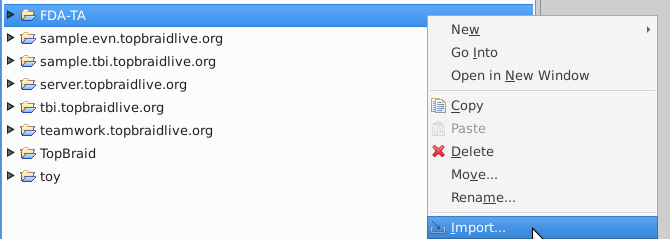
The files you need for this demo are available on the web. The following instructions load a data file from the web and save it in the project.
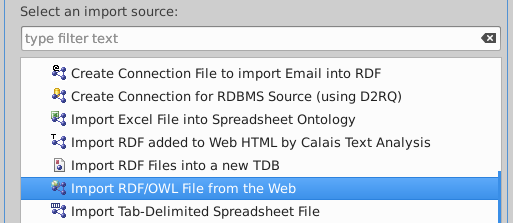
Select "Import RDF/OWL file from the Web"
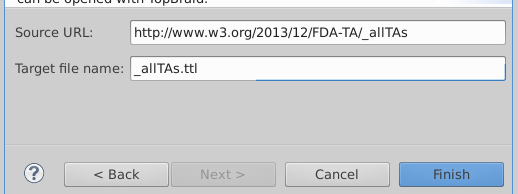
Enter "http://www.w3.org/2013/12/FDA-TA/_allTAs" in the Source URL and "_allTAs.ttl" for the Target file name
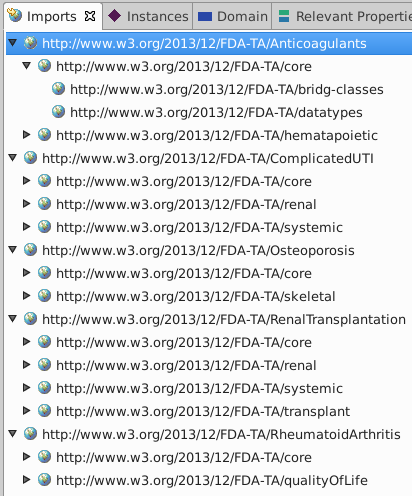
The Imports window will show the import tree:
- TBC made a copy of the file "_allTAs.ttl" and saved it in the project directory.
_allTAs.ttlimports several other parts of the ontology, e.g. "http://www.w3.org/2013/12/FDA-TA/Anticoagulants".- These imported resources are not saved locally; each time you load
_allTAs.ttl, TBC will re-import those resources with HTTP GET. - If you wish to use a web copy of
_allTAsyou can instead create a new file and import "http://www.w3.org/2013/12/FDA-TA/_allTAs" with the Imports pane.
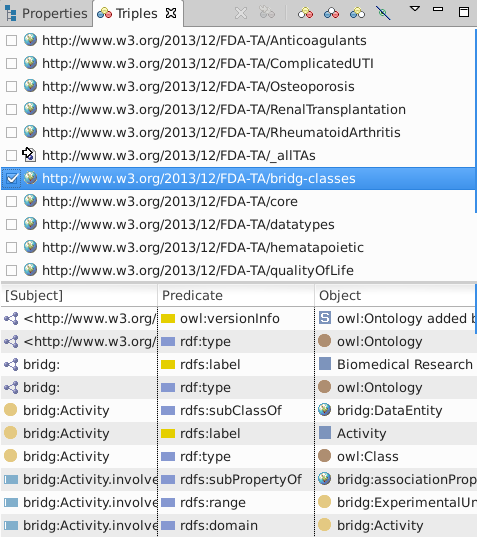
Review the loaded triples (Window·Show view·Triples):
Start class hierarchy with owl:Thing
This simplifies the rendered class hierarchy by hiding the syblings underneath the rdf:Resource root.
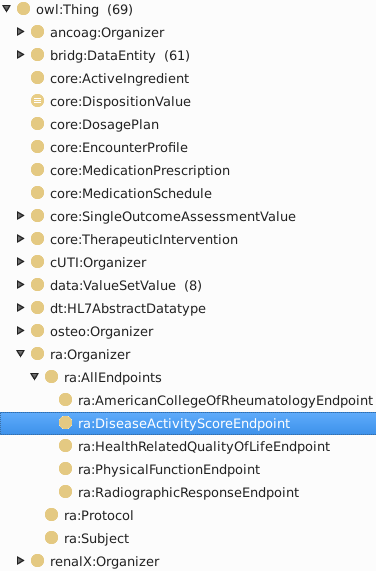
Expand the Rheumatoid Arthritis hierarchy
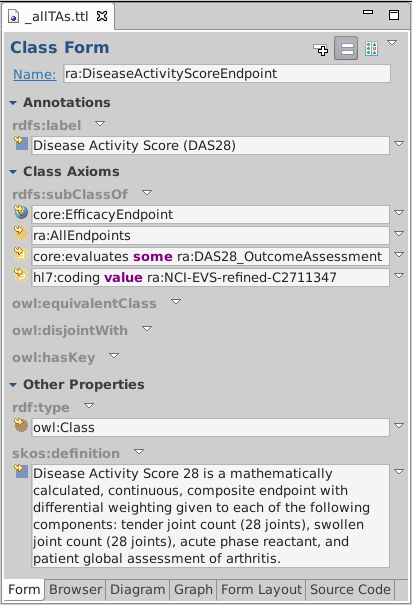
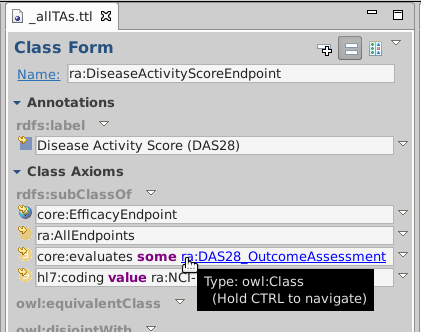
Double-click ra:DiseaseActivityScoreEndpoint to show class form
Use the arrows like a Web browser
to go back and forth through you traversal history.
TBC demo 2 - loading instance data
Follow the import instructions above to import and load "http://www.w3.org/2013/12/FDA-TA/tests/RenalTransplantation/subjects" into "subjects.ttl"
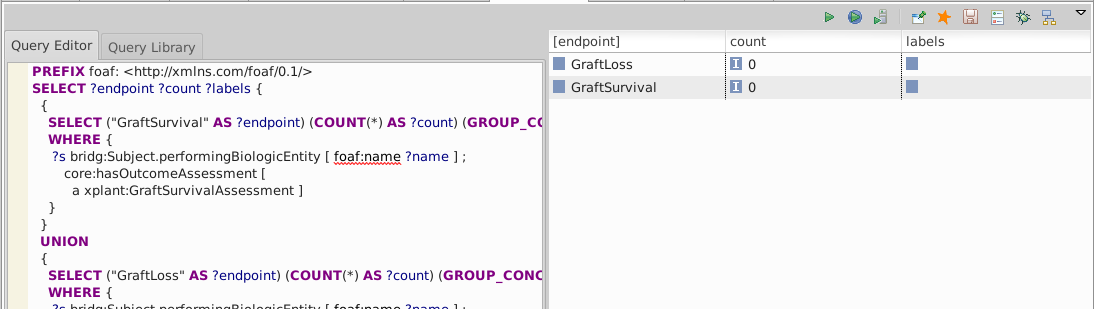
Execute an endpoints query over the loaded data
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
SELECT ?endpoint ?count ?labels {
{
SELECT ("GraftSurvival" AS ?endpoint) (COUNT(*) AS ?count) (GROUP_CONCAT(?name) AS ?labels)
WHERE {
?s bridg:Subject.performingBiologicEntity [ foaf:name ?name ] ;
core:hasOutcomeAssessment [
a xplant:GraftSurvivalAssessment ]
}
}
UNION
{
SELECT ("GraftLoss" AS ?endpoint) (COUNT(*) AS ?count) (GROUP_CONCAT(?name) AS ?labels)
WHERE {
?s bridg:Subject.performingBiologicEntity [ foaf:name ?name ] ;
core:hasOutcomeAssessment [
a xplant:GraftLossAssessment ]
}
}
}
The xplant:GraftSurvivalAssessment and xplant:GraftLossAssessment types are not asserted directly in the data so we expect counts of 0 GraftSurvival and GraftLoss:
Click the play button icon to run inferencing
View Inferences (Window·Show view·Inferences) to see the inferred triples
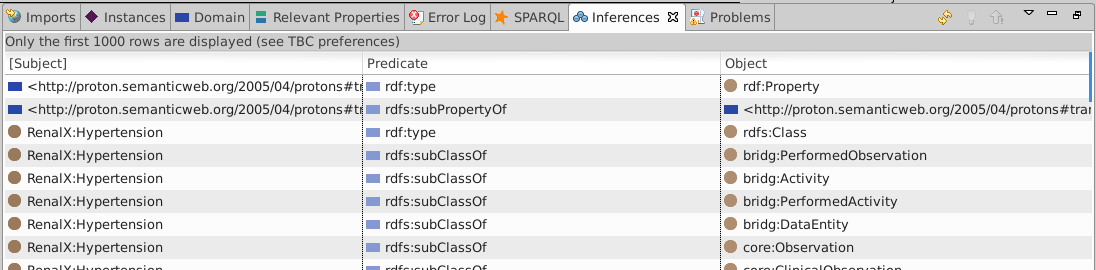 Somewhere in there are the
Somewhere in there are the xplant:GraftSurvivalAssessment and xplant:GraftLossAssessment type arcs required by the endpoint query.
Re-run the SPARQL query and see 1 result for each endpoint
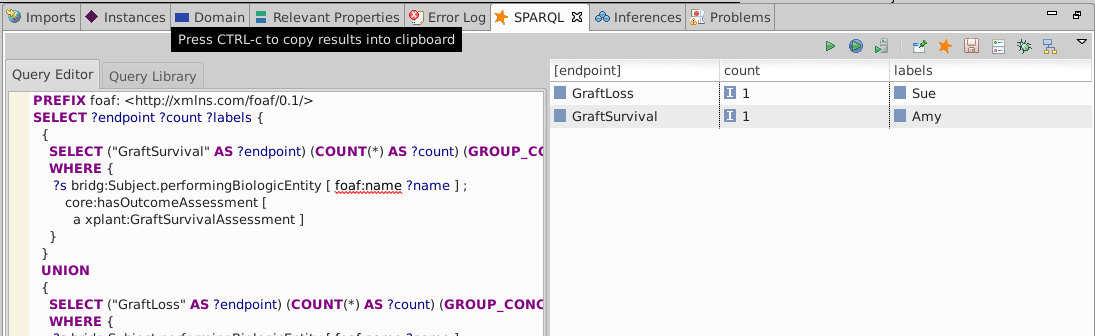 The loaded data has only three patients, including one with a graft survival (Amy) and one with a graft loss (Sue).
Had the data included more patients with clear loss or survival outcomes, their names would have been listed in the same row.
The loaded data has only three patients, including one with a graft survival (Amy) and one with a graft loss (Sue).
Had the data included more patients with clear loss or survival outcomes, their names would have been listed in the same row.
TBC demo 3 - sufficient reasoning
This demonstrates how OWL uses the properties of a node in a graph to discover the RDF type of that node.
Follow the import instructions above to import and load "https://www.w3.org/wiki/images/c/cc/Sufficient.ttl" into "Sufficient.ttl"
This imports a small ontology
xplant:GraftSurvivalAssessment
rdfs:subClassOf core:PositiveOutcome ;
owl:equivalentClass [
owl:onProperty core:hasResultValue ; owl:hasValue xplant:NormalFunctioningGraft
] .
data:someOutcome core:hasResultValue xplant:NormalFunctioningGraft .
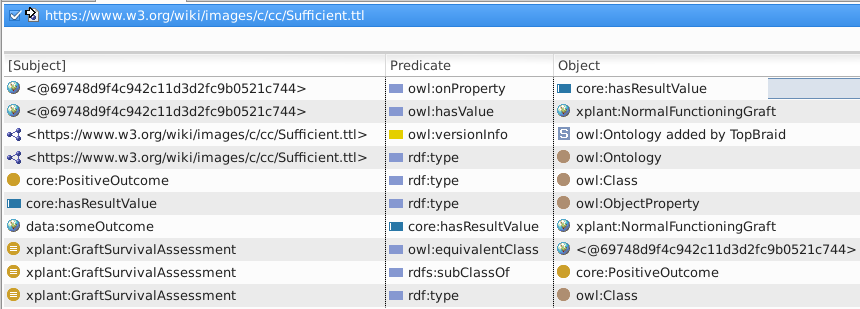
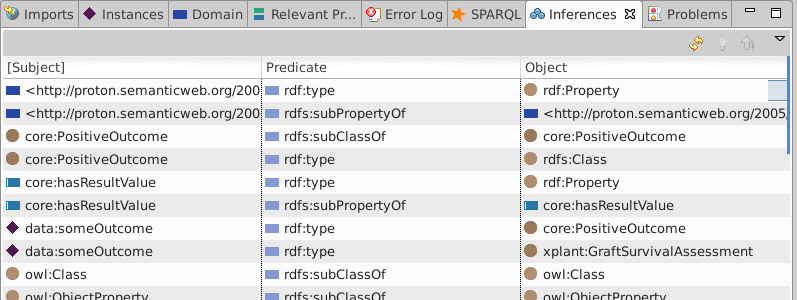
View Triples (Window·Show view·Inferences) and note the 4 type arcs
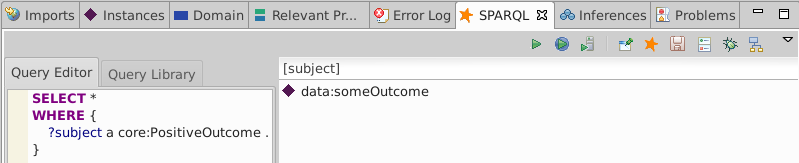
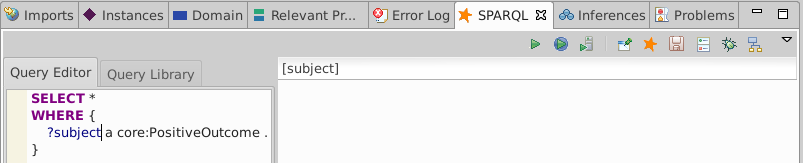
Execute a query for everything of type core:PositiveOutcome
SELECT *
WHERE {
?subject a core:PositiveOutcome
}
The core:PositiveOutcome type is not asserted so we expect no solutions:
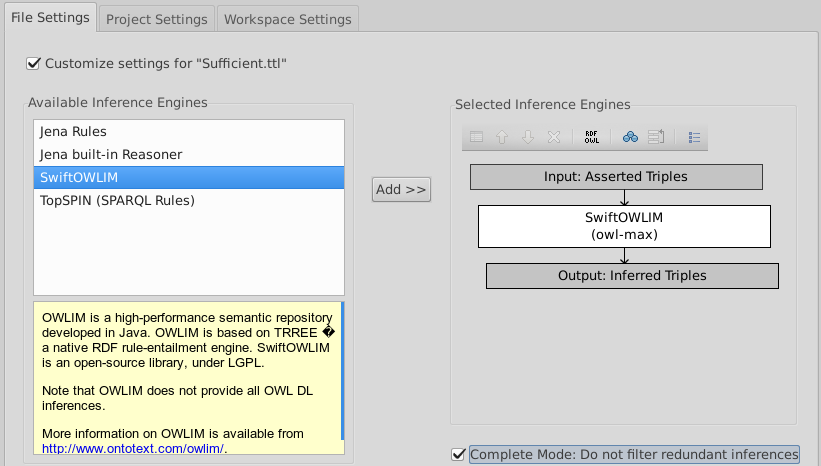
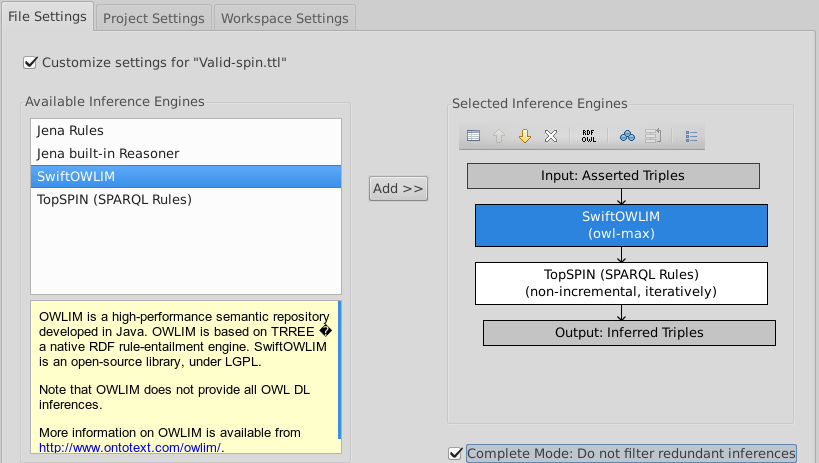
Change the inference from TopSPIN to OWLIM
- Select Inferences·Configure Inferencing...) from the menu.
File:TBC-FDA-TA-inference-TopSPIN.png
- Check 'Customize Setting for "Sufficient.ttl"'.
- Click the "TopSPIN (SPARQL Rules)" box on the right and the white 'X' above "Input: Asserted Triples".
- Double-click SwiftOWLIM on the left.
- Check "Complete Mode: Do not filter redundant inferences" at the bottom. You should now see:
Click the play button icon to run inferencing
View Inferences (Window·Show view·Inferences) to see the inferred triples
Note that one of the four inferred type triples in the image asserts data:someOutcome rdf:type core:PositiveOutcome.
If you did not check "Complete Mode" while configuring inferencing, you will only see four triples. These will not include transitive subclass inferencing, e.g. that data:someOutcome rdf:type core:PositiveOutcome.
Re-run the SPARQL query and see the type of data:someOutcome
TBC demo 4 - necessary reasoning
The demonstrates how OWL can infer properties for a graph node based on its type.
Follow the instructions for demo 3 above to import and load "https://www.w3.org/wiki/images/2/23/Necessary.ttl" into "Necessary.ttl"
This imports the small ontology
xplant:GraftCSARAssessment
rdfs:subClassOf
[ owl:onProperty core:hasResultValue ;
owl:hasValue xplant:NonFunctioningGraft ] .
data:someOutcome a xplant:GraftCSARAssessment .
Change the inference from TopSPIN to OWLIM as above
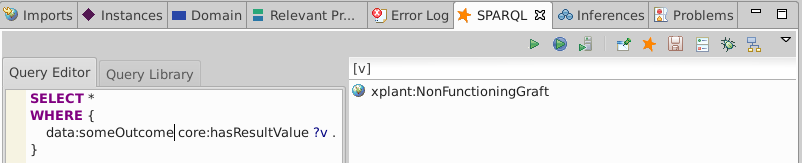
Execute a query for data:someOutcome's core:hasResultValue
As above, you will need to run inferencing first to see results.
SELECT *
WHERE {
data:someOutcome core:hasResultValue ?v .
}
Necessary and sufficient=
In general, owl:equivalentClass asserts that some pattern is sufficient to assert membership in a class and rdfs:subClassOf to assert that membership in a class implies that certain constraints are met.
The rdfs:subClassOf idiom may seem peculiar but it may be easy to visualize in terms of Venn diagrams, one representing the set of things in the world with a core:hasResultValue of xplant:NonFunctioningGraft and one which is the set xplant:GraftCSARAssessment.
Since xplant:GraftCSARAssessment is a subclass of the other set, we can see that every xplant:GraftCSARAssessment must core:hasResultValue of xplant:NonFunctioningGraft.
TBC demo 5 - validation
Follow the instructions for demo 3 above to import and load "https://www.w3.org/wiki/images/7/72/Valid-spin.ttl" into "Valid-spin.ttl"
This imports the ontology
xplant:GraftCSARAssessment
rdfs:subClassOf
[ owl:onProperty core:hasResultValue ; owl:hasValue xplant:NonFunctioningGraft ] .
[ owl:onProperty core:hasResultValue ; owl:cardinality 1 ] .
[ a owl:AllDifferent ;
owl:members (xplant:NonFunctioningGraft xplant:FullyFunctioningGraft) ] .
data:someOutcome a xplant:GraftCSARAssessment ;
core:hasResultValue xplant:FullyFunctioningGraft .
One might expect that data:someOutcome is obviously invalid because it has the wrong value for core:hasResultValue.
This would be the case in a DL reasoner (which would, perhaps surprisingly, require the owl:AllDifferent to know that xplant:FullyFunctioningGraft is not another name for xplant:NonFunctioningGraft), but an RL reasoner doesn't enforce cardinality.
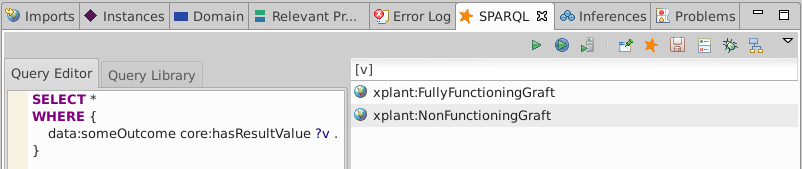
Looking at the inferred triples shows you that data:someOutcome core:hasResultValue xplant:NonFunctioningGraft..
Change the inference from TopSPIN to OWLIM as above
Re-add TopSPIN below SwiftOWLIM:
Execute a query for data:someOutcome's core:hasResultValue
As above, you will need to run inferencing first to see results.
SELECT *
WHERE {
data:someOutcome core:hasResultValue ?v .
}
We see that data:someOutcome has two core:hasResultValue. An OWL DL reasoner would flag this as an error because of the [ owl:onProperty core:hasResultValue ; owl:cardinality 1 ] constraint.
View Problems (Window·Show view·Inferences) and refresh the problem list
In order to enforce this cardinality constraint, we can use a SPIN rule.
Note that data:someOutcome is of type xplant:GraftCSARAssessment.
Adding a constraint to xplant:GraftCSARAssessment will allow us to make sure there's only ever one core:hasResultValue and that it's value is xplant:NonFunctioningGraft