Implementability requires:
Clinical Observation Interoperability requires:
:subjectsPostOpDay3SCr a renal:SerumCreatinineObservation ; core:hasObservationTime "2013-07-08T14:22:00Z"^^xsd:dateTime ; core:hasResultValue [ data:value 3.4 ; data:units ucum:mg_dL ]. :subjectsPostOpDay3GFR a renal:GfrFlowRateObservation ; core:hasObservationTime "2013-07-08T14:50:00"^^xsd:dateTime ; core:hasResultValue [ data:value 12.0 ; data:units ucum:mL-per-minute ].
:subjectsKidneyCSAR1 a RenalX:KidneyGraftCSARAssessment ; core:hasObservationTime "2013-07-07T19:00:00Z"^^xsd:dateTime ; core:hasObservation :subjectsPostOpDay3SCr , :subjectsPostOpDay3GFR ; core:hasResultValue xplant:NonFunctioningGraft . :subjectsRenalBiopsy1 a renal:RenalBiopsy ; core:hasObservationTime "2013-07-08T15:35:00Z"^^xsd:dateTime ; core:isJustifiedBy :subjectsKidneyCSAR1 ; core:hasLabReport :subjectsRenalBiopsy1report . :subjectsRenalBiopsy1report a renal:RenalBiopsyReport ; core:hasObservationTime "2013-07-08T16:10:00Z"^^xsd:dateTime ; core:hasClassification xplant:TCellMediatedRejection ; core:hasPathologyFinding xplant:BanffIII . :subjectsKidneyBPAR a RenalX:KidneyGraftOutcomeAssessment ; core:hasObservationTime "2013-07-08T16:15:00Z"^^xsd:dateTime ; core:beforeIntervention :subjectsOutpatientGFR23 , :subjectsGFRpreOp ; core:prescription :subjectOnImmunosuppressantB2 ; # core:hasIntervention … core:afterIntervention :subjectsRenalBiopsy1report ; # different type! core:hasResultValue xplant:NonFunctioningGraft .
see also intro slide 7,10,28
CSAR: Clinically Suspected Acute Rejection
BPAR: Biopsy-Proven Acute Rejection
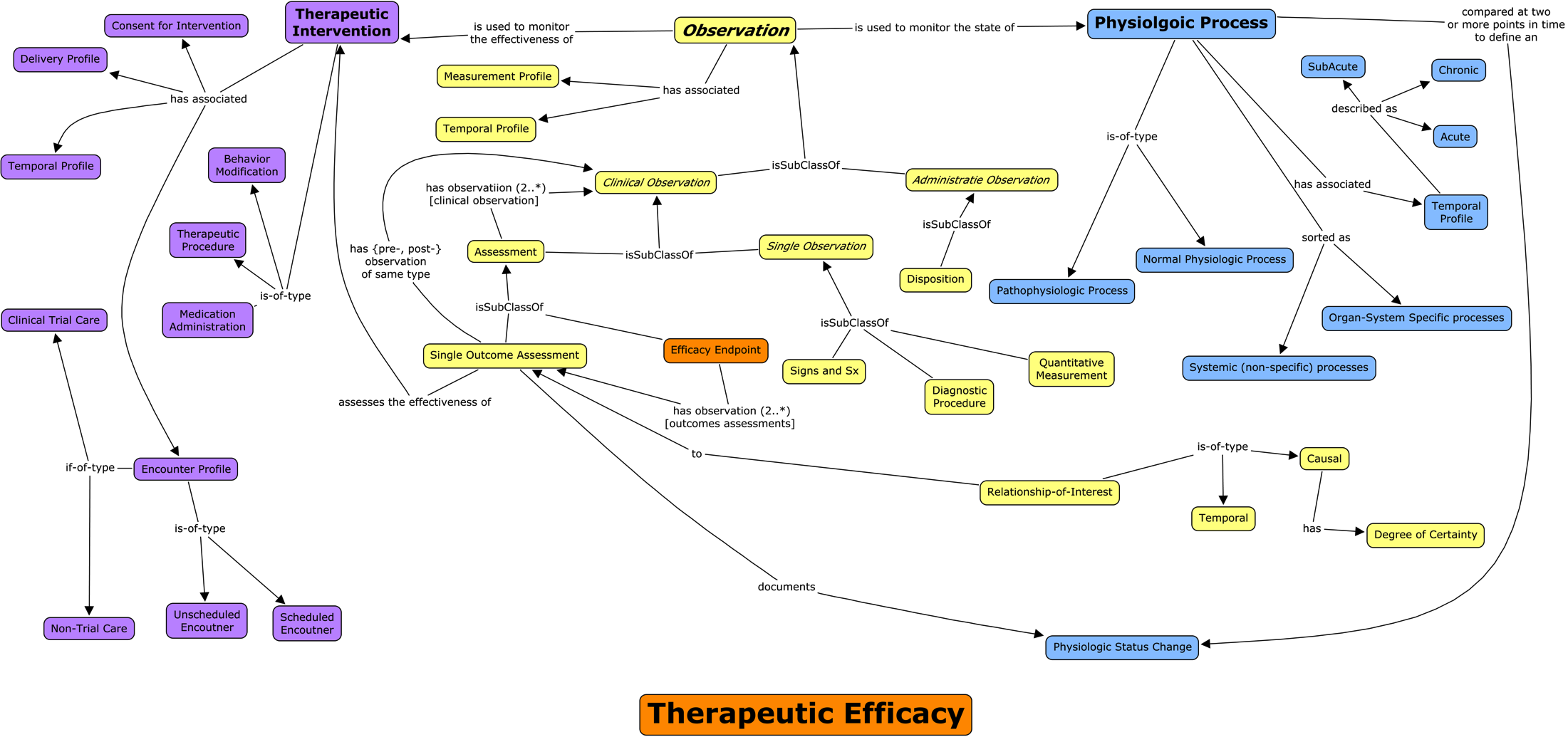
Concept maps are a human-intuitive way to communicate structures and relationships.
We need machines to work-with and enforce that structure.
Success metric: minimize implementation guidelines because the information is all in schema:

# Assessments are observations about other observations. core:Assessment rdfs:subClassOf core:ClinicalObservation , [ owl:onProperty core:hasObservation ; owl:minCardinality 1 ] . # Differentiate observations before and after an intervention. core:beforeIntervention rdfs:subPropertyOf core:hasObservation . core:afterIntervention rdfs:subPropertyOf core:hasObservation . # Specialize assessment to include before and after observations. core:SingleOutcomeAssessment rdfs:subClassOf core:Assessment , [ owl:onProperty core:beforeIntervention ; owl:minCardinality 1 ] , [ owl:onProperty core:hasIntervention ; owl:minCardinality 1 ] , [ owl:onProperty core:afterIntervention ; owl:minCardinality 1 ] . # A kidney graft outcome assessment is based observations of graft viability. renalX:KidneyGraftOutcomeAssessment ; rdfs:subClassOf core:FunctionOutcomeAssessment , [ owl:onProperty core:hasResultValue ; owl:allValuesFrom xplant:GraftViability ] .
see also intro slide 7, 29, 33
Comes from BRIDG and the meta-model. (intro slide 15)

Q: Is this a model to capture the results of valid protocol, or a way to generate valid protocols?
Should the models be detailed and restrictive enough to replace much of the existing protocols?
What alternatives did we have to BRIDG?
These don't capture:
If A and B share a semantic model, code can be written to translate between them.
If A and B also share a representation, no code is necessary.
Technically, "representation" should be called the "data model" but many things are called "model".
RDF has a few formats specialized to different purposes, but combining data is defined (and trivial) for all of them.
:SerumCreatinineLevel a owl:Class ; rdfs:subClassOf [ owl:onProperty bridg:resultIn ; owl:allValuesFrom bridg:PerformedClinicalResult ], [ owl:onProperty bridg:resultIn ; owl:cardinality 1 ] .
:GraftBPARAssessment a owl:Class ; rdfs:subClassOf core:NegativeOutcome ; owl:equivalentClass [ a owl:Class ; owl:intersectionOf ( [ a owl:Restriction ; owl:onProperty core:afterIntervention ; owl:someValuesFrom [ a owl:Restriction ; owl:onProperty core:hasPathologyFinding ; owl:hasValue :BanffIII ] ] [ a owl:Restriction ; owl:onProperty core:hasResultValue ; owl:hasValue :NonFunctioningGraft ] ) ] .
<RenalTransplantation> a owl:Ontology ; owl:imports <core> , <renal> , <transplant> .
see also intro slide 16
see also intro slide 20
*demonstrate FDA-TA directory*
*demonstrate TBC hierarchical view*
If it isn't clear where to write some facet down (e.g. TA-specific demographics, causality, diagnosis factors) the submitter will:
Results

Load the TA ontologies into Top Braid Composer see instructions in the wiki
imports
triples
class hierarchy
inheritance diagram
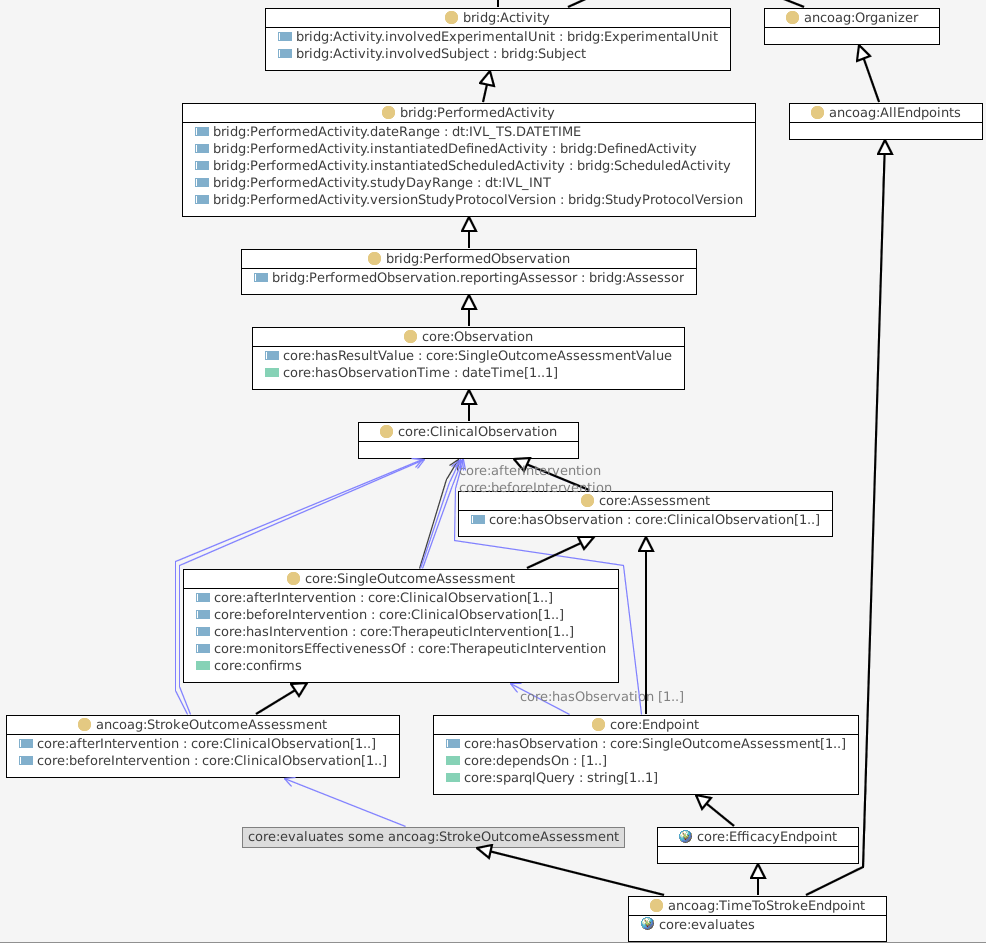
ancoag:StrokeOutcomeAssessment looks for a stroke (after some intervention).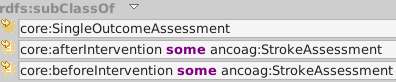
ancoag:StrokeAssessment is hemorrhagic, ischemic or uncertain.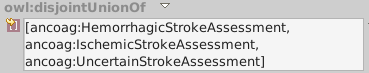
ancoag:IschemicStrokeAssessment is diagnosed by one of four observations.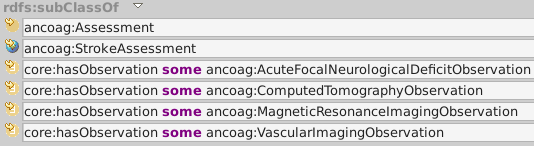
BRIDG + core provides a canvas to capture clinical trial exposures, observations and assessments in a structured way.
As described in the Machine-readable Structure slide, TA ontologies have to follow a rigid structure of endpoints measuring outcome assessments, which are in turn based on observations and assessments.
A purpose-built .TA (pronounced "dot T A") language has a grammar which follows (enforces) this structure.
Most errors one can make in a TA ontology document are prohibited by the grammar. intro slide 17
TA: RheumatoidArthritis IMPORT: "qualityOfLife.tapp" AS: qol: MEDHISTORY: OralCorticosteroid OralDisease-modifyingAntirheumaticDrug TNFInhibitorsBiologics NonTNFInhibitorBiologics MTX CONCOMITANTS: OralCorticosteroid OralDisease-modifyingAntirheumaticDrug TNFInhibitorsBiologics NonTNFInhibitorBiologics MTX MEDICATION: OralCorticosteroid @"Oral corticosteroids" COVARIATES: RheumatoidFactorObservation Anti-cyclicCitrullinatedPeptideObservation QUANT: RheumatoidFactorObservation @"Rheumatoid Factor" QUANT: Anti-cyclicCitrullinatedPeptideObservation @"Anti-CCP" EFFICACY: DiseaseActivityScoreEndpoint @"Disease Activity Score (DAS28)" DAS28_OutcomeAssessment({DAS28_Assessment}) ASSESSMENT: DAS28_Assessment @"Disease Activity Score (DAS28)" [0 10] SwollenJointCountObservation PatientGlobalAssessmentOfDiseaseActivityObservation QUANT: SwollenJointCountObservation @"Swollen joint count" [0 66] SSX: PatientGlobalAssessmentOfDiseaseActivityObservation @"Visual Analog Scale (VAS) for Patient Global Assessment"
Simplified development + factoring of shared libraries enables concurrent development.
Standard column headers provide:
skos:Definition for the concept.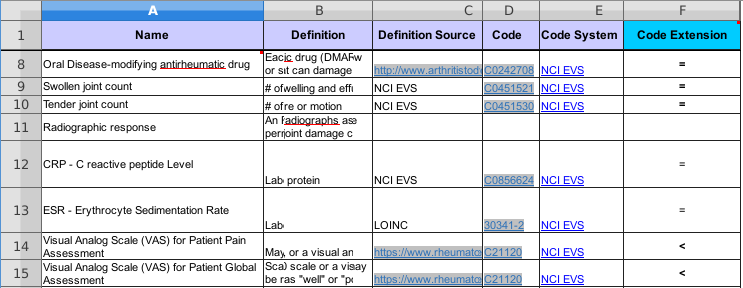
HL7 started with the UML and added hundreds of extensions, requiring in-house maintenance of custom tools.
OWL now captures most of these extensions.
Basing on RDF and OWL means there are 10s of reasoners and maybe 50-100 SPARQL engines.
SELECT ?outcomeType ?dose
(AVG(?endpoint-time) AS ?rate)
WHERE {
# drug of interest
?adminDrug dt:CD.displayName "Upsidasium" ;
… codingSystem … .
# subjects in studies about drug
?arm :studySubject ?adminDrug ;
:studyParticipation ?subject .
# demographic selection
?subject :taxon ncbitax:9606 .
# outcomes assessing prescription performance
?outcome :intervention ?p ;
:value [ a ?outcomeType ] .
# ... of that drug on that subject (participation)
?p a :Prescription ;
:involvedSubject ?subject ;
:medication ?adminDrug .
} ORDER BY ?dose see also intro slide 8
Outcomes across one study:
sparql -d study1.ttl endpoints.rq -8
Outcomes across two studies:
sparql -d study1.ttl -d study2.ttl endpoints.rq -8
The query
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
SELECT ?endpoint ?count ?labels {
{
SELECT ("GraftSurvival" AS ?endpoint) (COUNT(*) AS ?count) (GROUP_CONCAT(?name) AS ?labels)
WHERE {
?s bridg:Subject.performingBiologicEntity [ foaf:name ?name ] ;
core:hasOutcomeAssessment [
a xplant:GraftSurvivalAssessment ]
}
}
UNION
{
SELECT ("GraftLoss" AS ?endpoint) (COUNT(*) AS ?count) (GROUP_CONCAT(?name) AS ?labels)
WHERE {
?s bridg:Subject.performingBiologicEntity [ foaf:name ?name ] ;
core:hasOutcomeAssessment [
a xplant:GraftLossAssessment ]
}
}
}
Clinical observations queriable:
?arm :studySubject ?adminDrug
SELECT ?outcomeType ?dose
(AVG(?endpoint-time) AS ?rate)
WHERE {
# drug of interest
?adminDrug dt:CD.displayName "Upsidasium" ;
… codingSystem … .
# subjects in studies about drug
?arm :studySubject ?adminDrug ;
:studyParticipation ?subject .
# demographic selection
?subject :taxon ncbitax:7609 .
# outcomes assessing prescription performance
?outcome :intervention ?p ;
:value [ a ?outcomeType ] .
# ... of that drug on that subject (participation)
?p a :Prescription ;
:involvedSubject ?subject ;
:medication ?adminDrug .
} ORDER BY ?dose
see also intro slide 20
Reusing other ontologies enables sharing of queries and applications (minor effect) and teaching and comprehension investment (major effect).
Experts in this field should recognize structures and ideally point out improvements.
Portable BRIDG Representation
:Observation ; owl:disjointUnionOf ( :AdministrativeObservation :ClinicalObservation ) ; rdfs:subClassOf bridg:PerformedObservation , [ owl:onProperty :hasObservationTime ; owl:cardinality 1 ] .
xplant:GraftSurvivalAssessment rdfs:subClassOf core:PositiveOutcome ; owl:equivalentClass [ owl:onProperty core:hasResultValue ; owl:hasValue xplant:NormalFunctioningGraft ] . data:someOutcome core:hasResultValue xplant:NormalFunctioningGraft .
try sufficient.ttl with OWLIM inferencing
xplant:GraftCSARAssessment rdfs:subClassOf [ owl:onProperty core:hasResultValue ; owl:hasValue xplant:NonFunctioningGraft ] . data:someOutcome a xplant:GraftCSARAssessment .
try necessary.ttl with OWLIM inferencing
see also intro slide 29
:_endpoint_GraftSurvivalAssessment rdfs:subClassOf core:InferrableDiagnosis , [ owl:onProperty core:hasDiagnosis ; # !! undefined (as of migration from hasOutome) owl:someValuesFrom [ owl:onProperty core:hasResultValue ; owl:hasValue :NormalFunctioningGraft ] ] ; # = core:hasIntervention some :TransplantProcedure # and core:hasOutcomeAssessment some ( core:OrganFunctionOutcomeAssessment # and core:hasResultValue value renal:ImprovedToNormal) owl:equivalentClass [ owl:intersectionOf ( [ owl:onProperty core:hasIntervention ; owl:someValuesFrom :TransplantProcedure ] [ owl:onProperty core:hasOutcomeAssessment ; owl:someValuesFrom [ # produces error classes in Protégé: # rdfs:subClassOf core:OrganFunctionOutcomeAssessment , # [ owl:onProperty core:hasResultValue ; owl:hasValue renal:ImprovedToNormal ] owl:intersectionOf ( core:FunctionOutcomeAssessment [ owl:onProperty core:hasResultValue ; owl:hasValue :ImprovedToNormal ] ) ] ] ) ] .
see also intro slide 29
CONSTRUCT {
?this core:hasDiagnosis ?diag .
?diag core:hasResultValue :NormalFunctioningGraft
} WHERE {
?this core:hasIntervention ?intervention .
?intervention a :TransplantProcedure .
?this core:hasOutcomeAssessment ?oa .
?oa a core:FunctionOutcomeAssessment .
?oa core:hasResultValue :ImprovedToNormal
}
see also intro slide 29-32
xplant:GraftCSARAssessment rdfs:subClassOf [ owl:onProperty core:hasResultValue ; owl:hasValue xplant:NonFunctioningGraft ] . [ owl:onProperty core:hasResultValue ; owl:cardinality 1 ] . [ a owl:AllDifferent ; owl:members (xplant:NonFunctioningGraft xplant:FullyFunctioningGraft) ] . data:someOutcome a xplant:GraftCSARAssessment ; core:hasResultValue xplant:FullyFunctioningGraft .
try valid-spin.ttl with OWLIM inferencing
Requires DL reasoner (RL will infer two core:hasResultValue arcs and not respect cardinality).xplant:GraftCSARAssessmentShape { core:hasResultValue ( xplant:NonFunctioningGraft ) }
try valid-spin.ttl with OWLIM followed by SPIN inferencing
Separate language compiles to SPARQL which can be used as a SPIN constraint on xplant:GraftCSARAssessment.The simplest validation is to interpret the ontology with a close world and a unique name assumption.
<http://a.example/foo> owl:differentFrom <http://b.example/bar> .
Such an enumeration must be exhaustive, including even typos, which is an unreallistic requirement. The opposite tactic, more appropriate for instance validation, is to assume that names are distinct unless they are asserted to be equivalent.
<http://a.example/foo> owl:sameAs <http://b.example/bar> .
Every Person has a mother.
That mother is a Person.
see also intro slide 34 and wiki article on validation.
GFR observation MUST have a code ofloinc:GFR:ArVRat:Pt:Ser_Plas:QnORloinc:GFR:ArVRat:Pt:Ser_Plas:Qn:Cystatin-based_formula
A kidney transplant MUST have conformant GFR observations for baseline and every N days.
A BPAR assessment MUST be based on a CSAR assessment and a biopsy report of BanfII or BanfIII.
The pre and post observations of an assessment MUST have the same type.
see also wiki article on validation and Dublin Core Application Profiles validation requirements
Most clinical systems use some variant of ISO 21090 "CD" for codes from controlled terminologies:
# Example instance data -- Sue's baseline measured in cerebral spinal fluid :labObs1234 # Rheumatoid Factor Observations are defined in the Rheumatoid Artheritis ontology a ra:RheumatoidFactorObservation ; # All observations have a time core:hasObservationTime "2013-07-07T19:02:00Z"^^xsd:dateTime ; # We reuse the hl7 schema to capture the convention of a code and a code system (à la ISO 21090) hl7:coding [ dt:CDCoding.code "14034-3" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.1" ; dt:CDCoding.displayName "Rheumatoid factor:Arbitrary Concentration:Point in time:Cerebral spinal fluid:Quantitative" ; dt:CDCoding.codeSystemName "LOINC" ] ; # Instance data will of course have results... core:hasResultValue [ data:value 65 ; data:units ucum:u_mL ].
The code system and the code create a unique identifier.
see also intro slide 20 and wiki article Coding Specificity
_:obsX bridg:PerformedObservationCode [ hl7:coding [ dt:CDCoding.code "69405-9" ; dt:CDCoding.codeSystem "2.16.840.1.113883.6.1" ; dt:CDCoding.codeSystemName "LOINC" ; dt:CDCoding.displayName "Glomerular filtration rate/1.73 sq M.predicted" ] ].
_:obsX bridg:PerformedObservationCode loinc:69405-9 .
with expected metadata:
loinc:48643-1 dt:CDCoding.codeSystemName "LOINC" ;
dt:CDCoding.displayName "Glomerular…k" .
_:obsX bridg:PerformedObservationCode loinc:GFR:ArVRat:Pt:Ser_Plas:Qn .
loinc:loinc:GFR:ArVRat:Pt:Ser_Plas:Qn
rdfs:subClassOf evs:Rheumatoid_factor ;
owl:equivalentClass [
owl:intersectionOf (
[ owl:onProperty dt:CDCoding.codeSystem ; owl:hasValue "2.16.840.1.113883.6.1" ]
[ owl:onProperty dt:CDCoding.code ; owl:hasValue "69405-9" ]
) ] .
see also wiki article Coding Specificity and example URL definition.
finding site, method) to attach to measurement concepts (GFR measurement).
How do we constraint the data in a kidney transplant trial submission?
69405-9 OR 50210-4loinc:GFR:ArVRat:Pt:Ser_Plas:Qn OR
loinc:GFR:ArVRat:Pt:Ser_Plas:Qn:Cystatin-based_formulaWhat speech act is most appropriate?
What granularity best reflects the specificity required for merging tests?
What best communicates the requires imposed during the approval process?
see also wiki articles Clinical compatibility and Mapping LOINC to SNOMED
Getting as much re-use as possible from models and data as science or practice changes.
see also intro slide 5

code everything
minimize:
cope gracefully with:
interoperate with EHRS:
Existing Models/Standards
Controlled Terminologies:
Therapeutic Efficacy is comprised of: