Application of Standard Semantic Web Services and Workflow Technologies in the SIMDAT Pharma Grid
Changtao Qu, and Falk Zimmermann
C&C Research Laboratories, NEC Europe Ltd.
Rathausallee 10, D-53757 Sankt
Augustin, Germany
Email: {qu, zimmermann}@ccrl-nece.de
Abstract
The SIMDAT Pharma Grid is an industry-oriented, semantics enabled Grid environment whose purpose, among others, is to intelligently assist Biologists in conducting in-silico experiments through automating discovery, selection, composition, and invocation process of bioinformatics data services and analysis services. In the first system architecture design and prototype implementation, we apply sets of standard Semantic Web services and workflow technologies, in particular OWL, OWL-S, UDDI, and XScufl/Freefluo, to ensure interoperability, reusability, and maintainability of the system. In this position paper, we report on our current experiences regarding the benefits and drawbacks of leveraging these standard technologies in bioinformatics Grid applications.
Keywords: Semantic Web services, Workflow, Grid, OWL, OWL-S, UDDI
1. Introduction
SIMDAT (Data Grids for Process and Product Development using Numerical Simulation and Knowledge Discovery) is an EU/IST FP6 project (http://www.simdat.org) which aims at developing generic Grid technologies for the solution of complex application problems in four industrial application sectors: Automotive, Pharma, Aerospace and Meteorology. In the SIMDAT Pharma Grid, our focus is on enhancing an industrial bioinformatics data integration platform: SRS (Sequence Retrieval System, http://www.lionbioscience.com/srs) with a WS-I (further OGSA) compliant Grid architecture. Basically, we concentrate on the implementation of several advanced functionalities such as distributed data repository access, administration of virtual organization, workflow, knowledge discovery and data mining.
In this position paper, we mainly report on our current experiences of applying standard SWS (Semantic Web Services) and workflow technologies, in particular OWL, OWL-S, UDDI, and XScufl/Freefluo (http://freefluo.sourceforge..net/), in order to implement the semantic broker in the SIMDAT Pharma Grid. The goal of the semantic broker is to intelligently assist Biologists in conducting in-silico experiments through automating SRS service discovery, selection, composition, and invocation process.
2. Semantic Broker in the SIMDAT Pharma Grid
Until January 2005, SRS has over 300 installations around the world, which accommodate overall 1000+ SRS services. These services can be divided into two categories: SRS data services, which can answer a user’s query based on sets of biological databanks; and SRS analysis services, which can perform specific types of computational analyses against user-submitted biological data. In order to efficiently conduct in-silico experiments based on the SRS platform, a user is expected to have both a general knowledge about available SRS installations, and an in-depth understanding of many different SRS services (e.g., functionalities, QoS features, security requirements, etc.) and their relationships to each other. While these “prerequisite” knowledge may be at the heart of many Bioinformaticians, the majority of novice SRS users like Biologists, who have clear ideas about the in-silico experiment procedure but sometimes lack knowledge of SRS, may be a bit overloaded. In order to shield these users from the “prerequisite” SRS knowledge, in SIMDAT Pharma Grid we propose to use the semantic broker to reduce the user’s interaction with SRS services by means of automating SRS service discovery, selection, composition, and invocation process. In figure 1 we illustrate in part the first system architecture design of the SIMDAT Pharma Grid which is centred upon the semantic broker. The three kernel enabling technologies here are ontology, SWS, and workflow.
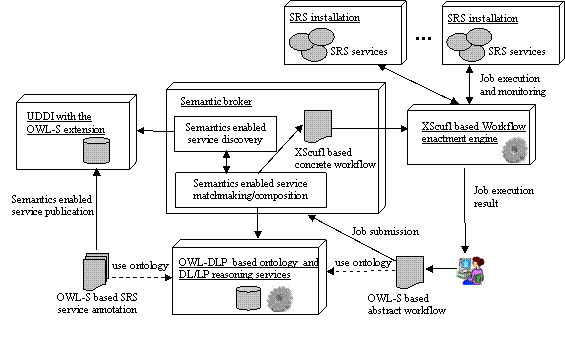 |
Figure 1. System architecture design of the SIMDAT Pharma Grid
3. OWL-DLP based Ontology and DL/LP Reasoning Services
The Ontology is the cornerstone enabling technology of the SIMDAT Pharma Grid. In the bioinformatics community, we can find a great variety of ontologies. However, as most of popular bioinformatics ontologies such as GO (Gene Ontology, http://www.geneontology.org/), SO (Sequence Ontology, http://song.sourceforge.net/), and OBO (Open Biological Ontologies, http://obo.sourceforge.net/) are simply DAGs-structured, controlled vocabularies lacking in formal logic descriptions, it is often very difficult to develop interoperable, reusable, and maintainable, application-specific ontologies based on them, which are evolving very fast. Without the formal logic support, it is also rather difficult, if not impossible, to implement advanced ontology reasoning services in order to support some more efficient usages of ontologies, e.g., intelligent ontology navigation, complex rule interpretation, etc. Moreover, the current status of bioinformatics ontologies has also misled “best practices” for developing semantics enabled bioinformatics applications, most of which are merely limited to using controlled vocabularies to annotate biological data within various databases and then leverage back-end specific query mechanisms to enable “semantic features” of the system. Although some pioneer bioinformatics ontology initiatives and applications such as GONG (Gene Ontology Next Generation, http://gong.man.ac.uk/) and TAMBIS (http://imgproj.cs..man.ac.uk/tambis/) tried hard to change this situation, most of them got frustrated by immaturity and instability of ontology standards in the past years.
Similar to myGrid (http://www.mygrid.org.uk/), one of our major design objectives in the SIMDAT Pharma Grid is to introduce some proven best practices popular in the SW (Semantic Web) community into the bioinformatics system design to develop interoperable, reusable, and maintainable semantics enabled bioinformatics applications. As currently most of SWS standards are relatively mature and stable, we intend to adopt a standard oriented design approach, taking system’s interoperability, reusability, and maintainability as the highest priority.
First of all, we adopt the W3C ontology standard OWL to develop application specific ontologies, which are essentially the subsets of some popular bioinformatics ontologies such as GO and SO but newly provided with the formal logic representation.. After having initially identified that OWL-DL can satisfy the knowledge modelling requirements regarding the SRS service annotation, we further restricted OWL-DL to its OWL-DLP subset with the intention of leveraging LP (Logic Programs) reasoning services in parallel to DL (Description Logic) ones. This is mostly motivated by the fact that LP reasoners have been well researched in the past years and we have got an excellent LP reasoner OntoBroker (http://www.ontoprise.de/) in SIMDAT, and also because we intend to retain the possible interoperability of the system with WSML-Core/WSMO. As we have cleanly separate the “external” ontology representation (OWL-DLP) from its “internal” knowledge model in reasoners (either LP or DL), we expect to seamlessly integrate both DL and LP reasoner in the SIMDAT Pharma Grid.
Second, we adopt a distributed and modularized ontology structure which is one of the key best practices in the SW community, but not yet fully acknowledged in the semantics enabled bioinformatics system design. In the SIMDAT Pharma Grid, we clearly differentiate between three ontology sets: SRS service ontology, which is used to describe SRS services classification, SRS data sources, and some general SRS service features such as QoS, IPR, etc.; SRS domain ontology, which is used to represent the domain knowledge based on popular bioinformatics ontologies; and the service annotation ontology, i.e. OWL-S. Besides these principal ontologies, we also plan to reuse some cross-domain ontologies such as vCard, Dublin Core, DAML security and privacy ontology (http://www.daml..org/services/owl-s/security.html), etc., with the purpose of further improving interoperability and reusability of our application specific ontologies. As an immediate benefit of such a distributed and modularized ontology structure, the SIMDAT Pharma ontologies can independently be developed and maintained by different project partners. They can also easily be distributed into different repositories independent of specific ontology usages and reasoning services.
Third, we implement ontology reasoning services in the SIMDAT Pharma Grid as standalone Web services, which can provide the reasoning support purely based on standard inferring operations such as subsumption, consistency checking, etc., but are independent of diverse ontology usages like the ontology validation, intelligent ontology navigation, ontology based service matchmaking, etc. At the first stage, the reasoning services are planned to be implemented in parallel using two types of reasoners: the DL reasoner Racer, and the LP/F-Logic reasoner OntoBroker. In the process of the SIMDAT project, we will evaluate and compare the performance of these two types of reasoners against large ontology sets.
Lessons learnt. Our demo ontologies are developed using Protégé 3.1 beta with OWL 2.0 plugin, which can efficiently facilitate the OWL ontology development process. However, we are aware that OWL 1.0 (Feb. 2004) itself still has some drawbacks, e.g., non-UNA makes the explicit disjoinness a “must” but the use of owl:disjointWith is rather inconvenient; owl:import is less efficient when importing large ontology sets, etc. With respect to reasoners, Racer is rather easy to use through a HTTP based DIG interface (there is also a file and TCP interface) for reasoning with OWL, but it still does not support full OWL-DL (e.g., inconsistent usage of UNA at the class level and instance level). OntoBroker is supposed to be able to provide strong reasoning support, however LP reasoner is deemed to be less native while reasoning with OWL.
4. OWL-S based SRS Service Annotation and UDDI based Service Publication
In the SIMDAT Pharma Grid, each SRS services are individually annotated based on the W3C OWL-S 1.1, and then published into a private UDDI registry with the OWL-S extension. At the first stage, we mainly focus on the OWL-S service profile and service grounding annotation for the SRS service discovery and invocation, but leave most of complex service model annotations such as “pre-condition”, “result”, as well as complicated control flow and data flow unaddressed. This is mainly because that most of open source APIs, editors, and rule languages (e.g., SWRL) are currently not yet full-fledged enough to support the complex OWL-S service model annotation, and also because we intend to simplify the first prototyping process.
In terms of the current SRS services annotation requirements, we initially choose following OWL-S service profile properties for the SRS services annotation (hasInput, hasOutput and hasParameter are actually defined in the OWL-S service model):
§ serviceName, textDescription: used to annotate general service information.
§ serviceClassification: used to annotate the service classification referring to the SRS service ontology.
§ serviceCategory: used to annotate the service category based on the NCBI Taxonomy (http://www.ncbi.nlm.nih.gov/).
§ serviceParameter: used to annotate service QoS (referring to the SRS service ontology), security (referring to the DAML security and privacy ontology), and SRS data sources (referring to the SRS service ontology).
§ hasInput, hasOutput, and hasParameter: used to annotate service inputs and outputs referring to the SRS domain ontology.
After the annotation process, each SRS service gets published into a UDDI registry via the OWL-S to UDDI mapping described in [1], i.e., each profile property is generally transformed into a UDDI tModel. As this mapping is not directly applicable to OWL-S 1.1 and UDDI 2.0, we fix the mapping according to the new standards, and further enrich the service bindingTemplate with the OWL-S service grounding. Due to the new UDDI 3.0 release (Feb. 2005), we plan to make further improvement on the mapping by introducing new UDDI 3.0 features such as the URI-based key, keyedreferenceGroups, catagoryBag in the bindingTemplate, etc., in order to enhance the efficiency of the UDDI based service publication and discovery.
Lessons learnt. The demo SRS services are annotated using SRI OWL-S editor alpha, which provides very nice support for the OWL-S 1.1 based service annotation. These annotations are then parsed through CMU OWL-S 1.1 API 0.1 beta, which is unfortunately not yet fully compatible with OWL-S 1.1, especially regarding the service model and service profile. However, CMU OWL-S 1.1 API seems to be working well with Protégé OWL API 2.0 and SRI OWL-S editor due to their common basis: HP Jena.
With regard to OWL-S 1.1 itself, some service profile properties still seem rather confusing, e.g., serviceClassification should apparently be defined as an ObjectProperty rather than a DatatypeProperty (xsd:anyURI) according to its usage described in the specification. With regard to the UDDI registry, we implement a UDDI 2.0 server using Apache JUDDI 0.9 rc3, Apache Tomcat 5.0, and MySQL database 4.1. The UDDI server seems to be working well with IBM UDDI4J 2.0.3 client-side API.
5. OWL-S based Abstract Workflow Description and XScufl based Workflow Enactment Engine
in-silico experiments in the SIMDAT Pharma Grid are described using the abstract workflow. “Abstract” in this context means that the workflow does not include the detailed binding information such as the service endpoint, invocation protocols, etc., to each of its operations. The SRS users describe each abstract workflow operation based on OWL-S, using the same ontology sets as for the SRS service annotation, to express their functional and non-functional requirements on each in-silico experiment step. As the current OWL-S 1.1 cannot directly be used to represent the workflow involving inter-services descriptions, we adopt another workflow language XScufl/Freefluo to “externally” describe the control flow and data flow between each workflow operations. Another option, which seems even more promising, is to extend the OWL-S service model to support the description of the workflow, i.e. taking a workflow as a kind of OWL-S composite services. As such an extension is rather critical and may damage our standard oriented design approach in SIMDAT, we do not go for this option at the first stage.
After the service matchmaking/composition process, the abstract workflow is interpreted as the executable concrete workflow, which is fully represented in XScufl and further fed into the XScufl based workflow enactment engine for execution.. In the SIMDAT Pharma Grid, the workflow enactment engine is a classical Grid run-time component, which is responsible for invoking SRS services and implementing various Grid run-time functionalities such as the data staging, service monitoring, fault management, event notification, etc. At the first stage, we plan to adopt Freefluo as the workflow enactment engine, which natively supports XScufl among other workflow languages.
Lessons learnt. We are still lacking a semantic workflow description language in the SWS community. XScufl is preferable in the Grid community because it is derived from BPEL and supported by the workflow editor Taverna (http://taverna.sourceforge.net/) and workflow enactment engine Freefluo. From a long-term point of view, the semantic workflow description language needs surely be drawn up by the SW or SWS community.
6. Semantics enabled Service Matchmaking and Composition
In the first prototype implementation, we mainly focus on matching each abstract workflow operation to SRS service instances based on the SRS service annotations, which are retrieved by the semantics enabled service discovery module from the UDDI registry. As we have not yet annotated the complex OWL-S service model, we can only expect to implement a simple service composition through reasoning with outputs and inputs of the required workflow operation by means of the backward chaining algorithm. In the next stage, we may exploit some more complex service compositions, provided some supporting technologies such as state machines, Petri nets and AI planning are well researched in the SW or SWS community..
At the first stage, we initially implement the semantics enabled service matchmaking based on the matching algorithm proposed in [2], which identifies four types of semantic “degree of match” between the “requirement concept” and “advertisement concept” (exact, plugin, subsume, fail). In terms of the practical usage of SRS services, following OWL-S service profile properties are matched according to their precedence orders, from the highest to the lowest.
serviceName -> serviceClassification -> serviceCategory -> serviceParameter(SRS Data Sources) -> serviceParameter(Security) -> serviceParameter(QoS) -> hasOutput-> hasInput
Furthermore, we plan to develop some service ranking algorithms based on the semantic “degree of match” of each properties through experimentally determining their weight values.
Lessons learnt. The semantics enabled service matchmaking is implemented using CMU OWL-S 1.1 API, Protégé OWL API 2.0 and Protégé API 3.1. We find that Protégé OWL API 2.0 is particularly handy while being used to query and manipulate OWL data models as well as to perform reasoning.
Our intent to exploit the complex service composition is however frustrated by the complexity of using OWL-S 1.1 and some “external” rule languages (e.g., SWRL) to annotate the service process. In addition, we are also aware that some supporting technologies such as OWL ontologies for state machines and Petri nets are still missing.
7. Conclusions and Future Work
Although there still exist some concerns about the stability of open standards as well as robustness and performance of open source products, our first system architecture design and prototype implementation of the SIMDAT Pharma Grid has initially shown several notable advantages of the standard oriented design approach, in particular interoperability, reusability and maintainability of the system, which, in our opinion, have not yet received enough attention in the bioinformatics community. From a long-term point of view, we are confident that such advantages will become increasingly obvious. In the next stage of the SIMDAT project, we will mainly focus on enhancing system functionalities to unleash the full power of open standards, e.g., exploiting the complex OWL-S service model annotation for the service matchmaking and composition, investigating system’s interoperability with the WSMO framework, etc.
8. Acknowledgements
We are grateful to all project partners at the SIMDAT Pharma Activity Consortium, in particular Chris Dodge from Lion Bioscience AG, Moritz Weiten and Martin Weindel from Ontoprise GmbH, and Kai Kumpf from Fraunhofer Institute for Algorithms and Scientific Computing.
References