
Marja-Riitta Koivunen
Annotea shared bookmarks can help researchers to organize their knowledge from different fields and sources. Annotea also helps groups of researchers to easily share their bookmarks and topics with selected other research groups. This document gives an example of how Annotea can be used to organize cancer related knowledge by using examples from chronic myelogenous leukemia (CML). The results are published as Annotea shared bookmarks and topics metadata.
CML is an uncommon blood cancer that is characterized by an overabundance of white blood cells and a missing piece of Philadelphia chromosome (22) caused by gene translocation between it and chromosome 9. It is a good example as there is a lot of information related to the disease, related genes, enzymes and treatments available on the Web.
In the following we describe a hypothetical scenario where our researcher Eric is using Annotea shared bookmarks and topics to organize some information available on the Web about CML. At first he creates some general topics and adds links to general articles of CML to them. Then he looks for more specific information about genes, pathways, enzymes, and ontologies. He also looks information about the medicines used for treating CML.
This scenario reflects the way the researchers can use Annotea shared bookmarks to create and modify their own views to lifescience information and share it with each other when much less is known than what we know about CML today. The Amaya user interface of Annotea shared bookmarks is still a prototype that can be considerably enhanced. Our aim is to get this implemented in all browsers either natively or through some plugins or proxies.
First our researcher, Eric, creates a couple of general topics by using Annotea. He locates the topic "Chronic Myelogenous Leukemia" under "Leukemia" as he thinks he might later add more information about other kinds of leukemias too and he wants to link that topic to other places or ontologies describing leukemia topic.
Eric also creates "Involved genes" topic as he knows genes are involved and he is interested in findind out more about how they affect this disease. Under CML he creates "General info" topic for some selected general articles and information about CML that he sees the general search engines, such as Google, returning when querying about the disease.

Figure 1: Creating a new topic by using the "Bookmarks" menu.
A topic is created by selecting "New topic" item from the "Bookmarks" menu as shown in Figure 1. It opens the "Topic properties" window presented in Figure 2. When the window opens it has most fields prefilled to help the user. Eric selects the right parent topic "Chronic Myelogenous Leukemia" and fills in the title of the topic to the "Title" field. Even though not required he also wants to fill the description field as it may help him or other researchers later.
Eric decides to create other topics later after making some bookmarks to generic papers and reading them to get ideas of kinds of topics he would like to use.
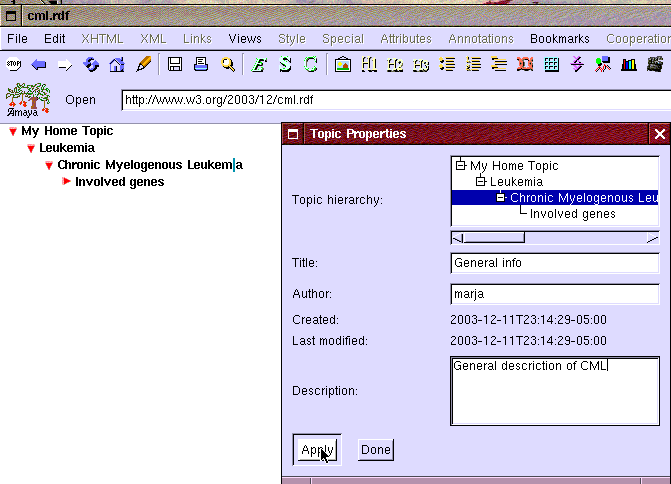
Figure 2: Creating new topic: General info under Chronic Myelogenous Leukemia
All the topics will have an unambiguous URI or URL address that is given automatically when the topics are published. The user interface is not showing the URI right now but the plan is to be able to do that in the future.
Eric looks for general articles of CML with Google by using search words "cml leukemia pubmed". He adds pubmed to the search words as he knows about pubmed database containing references to scientific articles. He receives the results shown in Figure 3 and opens the article "Chronic myelogenous leukemia in chronic phase.
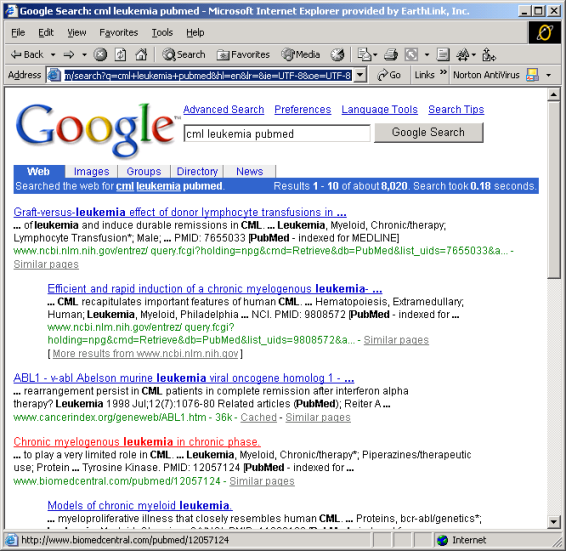
Figure 3: Searching Google with using keywords "cml leukemia pubmed".
Eric opens one of the articles named "Chronic myelogenous leukemia in chronic Phase" and makes a bookmark to this article by selecting "New bookmark" from the bookmark menu shown earlier in Figure 1. It opens the "Bookmark properties" window shown in Figure 4. Eric selects topic "General info" for the bookmark and clicks the apply button.
In addition, Eric wants to create another bookmark to the main PubMed database in case he needs to find other articles too. He opens the main PubMed page but before bookmarking it creates topics for "Search databases" and "Articles". Then he adds the bookmark the same way as earlier in Figure 4 by selecting "New bookmark" item from the menu.
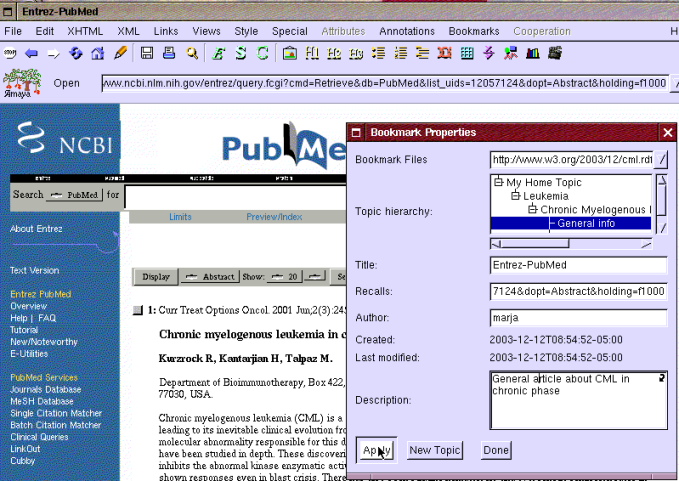
Figure 4: Creating a bookmark under topic: General info.
After creating a bookmark Eric realizes that the automatically created title "Entrez-PubMed" is not very descriptive. He opens the bookmark properties window by clicking the bookmark he just created with the right mouse button and changes the title to "Entrez-PubMed: CML in chronic phase" as shown in Figure 5. He does not remove "Entrez-PubMed" as he thinks that it might help him to know the source of the information later.

Figure 5: Change the title of a bookmark
Eric reads the abstract of the paper and finds out that the gene responsible for CML is called BCR-ABR. He does another search in PubMed and finds an article describing the structure of the gene "Structure of the Bcr-Abl oncoprotein oligogomerization domain." He bookmarks that under involved genes.
He also searches more information in PubMed database about STI571 molecule that can be used to treat CML (see Figure 6). He finds article "Imatinib mesylate therapy improves survival in patients with newly diagnosed Philadelphia chromosome-positive chronic myelogenous leukemia in the chronic phase.". He adds a topic "Treatment" and bookmarks the article under it.
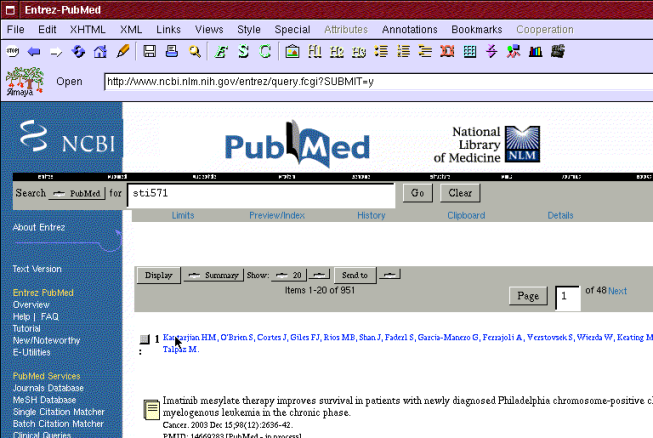
Figure 6: Search for STI571 molecule information in PubMed database
Eric is now getting interested in learning more about the genes and pathways that affect CML. He has heard about the cancer genome anatomy project (CGAP) so he finds that page and bookmarks it for later use under "Search DataBases/Genes" topic. Then he uses the gene finder tool for humans selecting keywords "bcr abl" that he found from some of the PubMed articles earlier. He gets the results shown in Figure 7. The last entry that does not have a symbol seems to be exactly what he is looking for.
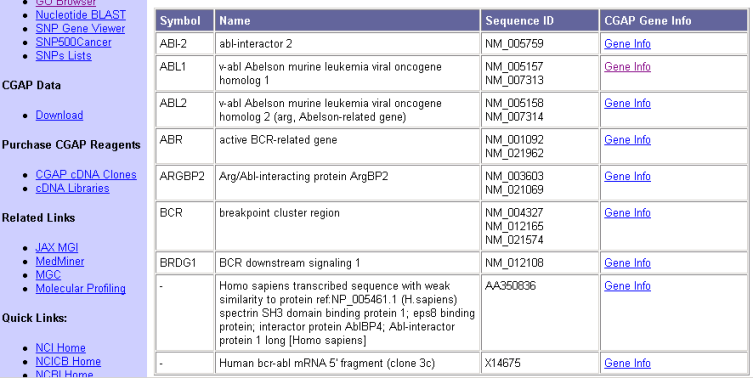
Figure 7: CGAP Gene finder results with keywords "bcr abl"
Eric follows the "Gene info" link in the "Human bcr-abl mRNA 5' fragment (clone 3c)" gene. The page contains the information presented in Figure 8.
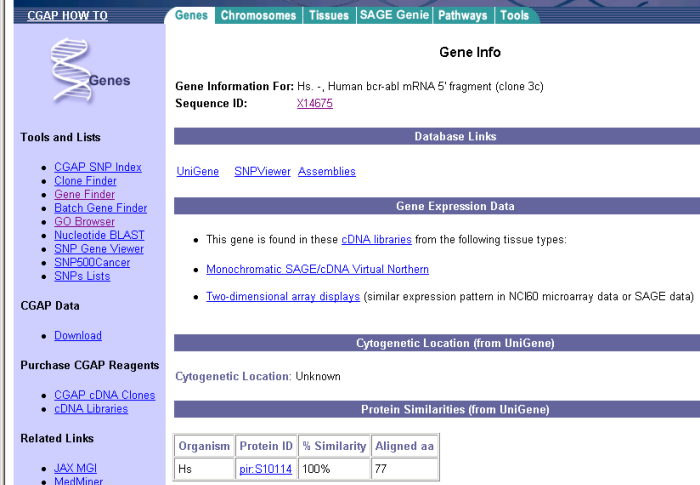
Figure 8: CGAP gene information for BCR-ABL gene.
Eric is still not sure if this is information of the same gene that the articles he found earlier discussed so he follows the link to the sequence ID X14675 information. This goes to a Nucleotide database. He looks the keywords in the gene data and they talk about Philadelphia chromosome, abl and bcr gene and tyrosine kinase. All these were also described in the papers and Eric is getting more confident this is the right gene.
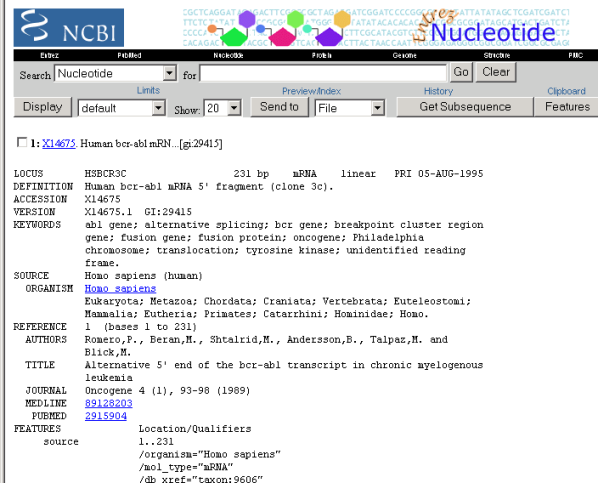
Figure 9: Gene with CGAP ID X14675
Eric bookmarks the CGAP page it under his "Involved genes" topic by using the "New bookmark" menu alternative and filling the information in the bookmark window presented in Figure 10.

Figure 10: bookmarking the gene info under "Involved genes" topic.
Eric is familiar with the Gene Ontology or GO database (http://www.geneontology.org/) and wants to do some gene related searches there to find good terminology to talk about the genes related to CML and also to find information about the genes from the databases using the GO terminology. As Eric already knows about the BCR-ABL gene he searches the database for "BCR-ABL" gene product information. But for some reason the database does not return that information. It does know about human BCR and ABL genes separately but not about the translocated gene. Eric decides to just make a bookmark to the human BCR gene in the GO database. Unfortunately the pages created by AmiGO are not valid according to standards and therefore are not presented properly by all browsers.
Eric hears from his friend Kate that LocusLink has good information about genes. He first searches for BCR-ABL gene and finds information about the BCR gene. He opens that by following the link "613" related to BCR and realizes that it seems to be more comprehensive information than what he retrieved from via AmiGO. So he happily bookmarks that too as shown in Figure 11.
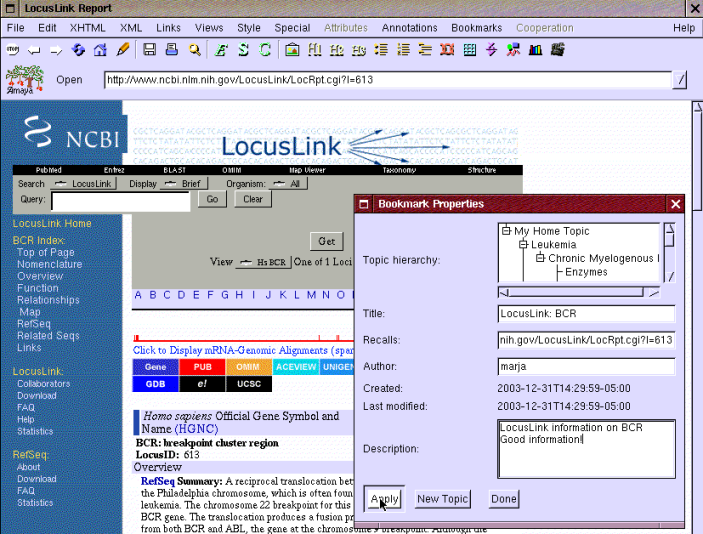
Figure 11: Bookmarking BCR gene in LocusLink
Eric looks for some GO terms related to tyrosine kinase activity that he may later want to create his own topics for and link to these terms from these topics. It is not yet implemented but in the near future the topic window will have a category "related topics" that lets Eric easily make these connections.
Eric searches pathways related to BCR-ABL gene by using the gene search provided by CGAP for BioCarta and KEGG databases. For some reason nothing can be found with "BCR-ABL" or even "BCR*" keywords. Similarly the Sequence ID X46175 for Human bcr-abl mRNA 5' fragment (clone 3c) does not retrieve any results.
Finally Eric tries a google search with "BCR-ABL" and "pathway" keywords. This returns "Inhibition of Cellular Proliferation by Gleevec" pathway. Eric makes inserts topic "pathways" to the bookmark file and inserts the found pathway under this topic. Unfortunately the biocarta active server pages seem not to be valid according to the standards and therefore can be only seen properly by selected browsers.
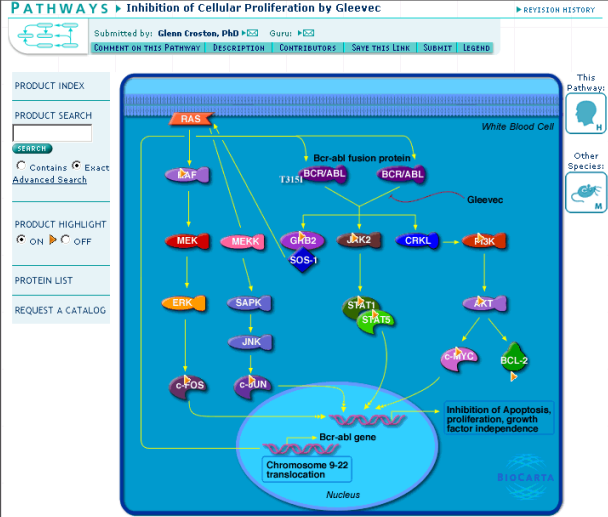
Figure 12: Pathway related to BCR-ABL gene and Gleevec inhibition
Another pathway related to BCR-ABL gene can be found by clicking the BCR/ABL object in Figure 12 and following the pathways link to a list of pathways related to BCR-ABL gene. The new pathway is named "Integring signaling pathway".
Eric has now gathered a basic set of CML related bookmarks and organized them under selected topics. Eric has stored the bookmarks and the topics to a selected URI http://www.w3.org/2003/12/cml.rdf, so that they are available for his co-workers (Note: the local URI's are going to be changed to global URI's in the next versions).
Eric's bookmark and topic information can now be used to find the bcr-abl related info that for some reason was hard to find by using the search engines of the specialized gene and pathway databases. The information can also be easily merged with other bookmark and topic information that Eric's co-workers have gathered about CML or related diseases.
Finally, it is possible to make connections from the topics to related topics and make searching the bookmark and other metadata easy for the researchers.

Figure 13: The bookmark file presented by Amaya
bookmark file: http://www.w3.org/2003/12/cml.rdf
Annotea home page: http://www.w3.org/2001/Annotea/
Marja-Riitta Koivunen $Date: 2003/12/31 21:47:24 $