Innovation in clinical research and bioinformatics has been severely hampered in part by the fragmented gathering and storing of data, reflecting the compartmentalization of medical science and practice. It has also been hampered by the programmatic necessity of keeping up with medical advances, which has led within every discipline to a multiplicity of special-purpose databases. Neither seamless integration nor simple extensibility of data stores is the norm. Yet to answer ad hoc questions cutting across disparate domains, clinical knowledge housed in isolated silos needs to be integrated. To make matters worse, typically clinical knowledge is expressed in ambiguous, idiosyncratic terminology. This is especially problematic for longitudinal patient data that can feasibly span multiple, geographically separated sources—pharmacies, local practices, group practices, and primary, secondary, and tertiary hospitals—and disciplines such as genetics, pathology, cardiology, etc. Without aid of a well-defined, standardized knowledge representation, the expense of ad hoc integration is formidable to impossible.
Semantic Web technology, and its various engineering specifications, seeks to remove some of these barriers. It combines a highly-distributable addressing and naming mechanism (Uniform Resource Identifiers: URIs) with a formal knowledge representation (RDF and OWL), a mechanism for rendering document dialects in this knowledge representation (GRDDL), and a common query language (SPARQL). The multifaceted nature of URIs alleviates some of the accessibility challenges associated with physically separated components. The common knowledge representation empowers domain experts with a language for capturing clinical terminology formally and with little ambiguity. Assertions can be added at a later point with no impact to the organization of physical storage and minimal impact on existing terminology. SPARQL provides a common query language for accessing assertions expressed in such terminology. Finally, GRDDL bridges gaps between messaging dialects (such as HL7 RIM) and more expressive terminologies (such as SNOMED-CT).
The Cleveland Clinic has been heavily involved in both developing and applying Semantic Web standards. The goal is to improve the Clinic’s ability to use patient data for generating new knowledge to improve future patient care through outcomes-based and longitudinal clinical research. In addition, expressiveness and versatility of formats being used has been leveraged to provide individual patients an appropriate terminology and accessible view of summary data.
In addition to the challenge of ad hoc querying complex, scattered, longitudinal patient data, an equal challenge to clinical research and bioinformatics is the infrastructure needed for ad hoc data collection. Thick-client solutions have proven cumbersome and aligned more closely with document repositories and databases than knowledge bases. Over the last 4 years, Cleveland Clinic has developed a representational methodology for bridging data collection, document management, and knowledge representation. The result is a unified content repository called SemanticDB. SemanticDB has been internally deployed for production on top of the open source XML & RDF content repository bundled with 4Suite and Mozilla’s Firefox with XForms extensions. The methodology is realized through a core set of terms that facilitate creation of a domain vocabulary (or domain model) such that instances of the vocabulary can in some substantial and important ways be managed automatically by the system (see Figure 1).
Through automated application of generated data transforms, patient records are stored and readily available as both uniform, structured markup and as RDF. The coordinated use of both representation languages affords a variety of powerful operations on the patient record content: form-based data entry, transformation to reporting formats, document validation, targeted inference, and querying. These operations can be dispatched on the patient record documents and RDF graphs over a uniform set of interfaces.
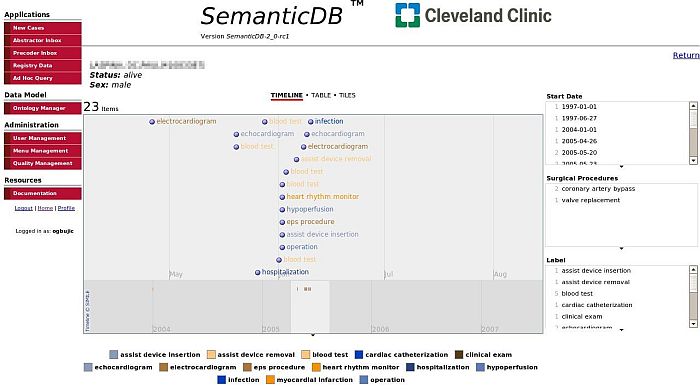
Figure 1: Screenshot of the SemanticDB Patient Record Portal (a larger version of the screenshot is also available)
This platform (illustrated in Figure 2) is to serve as the vehicle for the clinical research pipeline in the Department of Thoracic and Cardiovascular Surgery. It spans patient record abstraction and data entry via knowledge-generated input screens, work flow management, data quality management, identification of patient cohorts for study, and data export for statistical analysis. The dataset is a faithful RDF rendition of a set of some 195,000 patient records, all of which are managed in an instance of SemanticDB.
At the time of this writing, the patient record dataset has upwards of 54.2 million RDF assertions. The dataset is managed using the RDFLib open source RDF library and its MySQL adapter, which delegates physical storage to a centralized MySQL database. The MySQL database resides on an SGI Altix 350 super computer. In addition, stored SPARQL queries can be dispatched within a large distributed computing cluster.
Finally, the ability to infer new knowledge through the of ontology languages such as OWL, and rule languages such as Notation 3, affords a high-level of automation of most components typically associated with the management of data over their lifetime. A single description of a specific domain generates a detailed data dictionary, an XML schema, a formal ontology, and re-usable data transformations. These provide the foundational infrastructure for a SemanticDB instance.
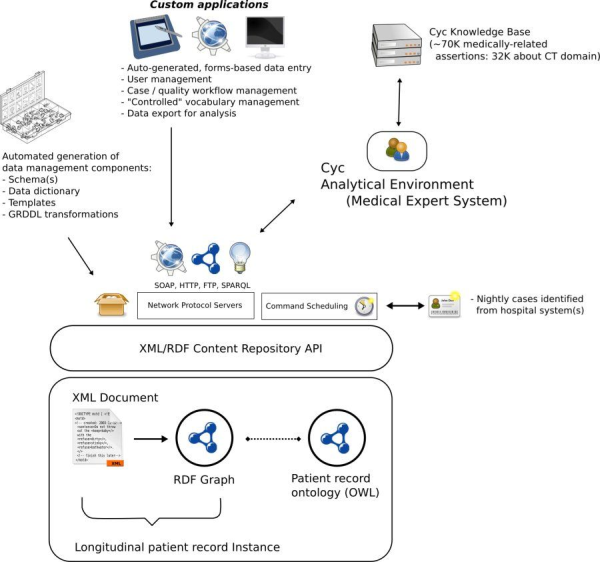
Figure 2: SemanticDB component architecture (a larger version of the image is also available)
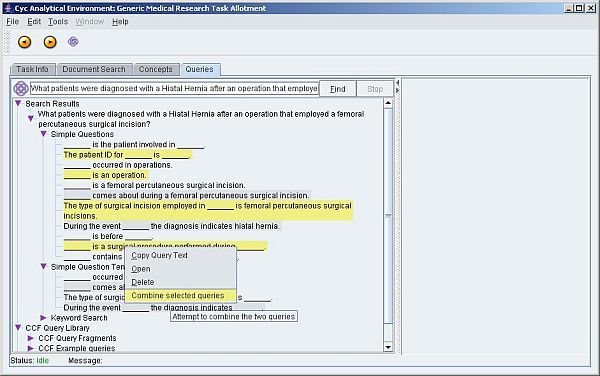
Figure 3: Forming a query from fragments in natural language (a larger version of the screenshot is also available)
During 2007, the bioinformatic team has collaborated with Cycorp Inc. to develop a state-of-the-art query interface that builds query fragments through natural language-driven interactions with a clinical investigator (see Figure 3). These query fragments are compiled into a sequence of optimized SPARQL query fragments that are evaluated against a remote SPARQL service.
The service supports an RDF dataset of cardiovascular procedures, with some additional assertions about infectious diseases (representing a disparate discipline). The translation is made possible by a carefully curated mapping from an exported OWL ontology of the dataset to the Cyc consensus ontology. This has resulted in a highly expressive common language for the domain of thoracic and cardiovascular surgery. The Cyc Analytic Environment (as the interface is called) will be the common entry point for investigative research on the RDF dataset.
A major difference between the user experience with SemanticDB and the previous interface to the relational technology-based Cardiovascular Information Registry (CVIR) that has accelerated adoption of Semantic Web technologies is the use of local terminology familiar within the domain rather than terms that are a consequence of the physical organization of the data. In addition, the model of the domain (expressed in OWL) is more amenable to extensions typically associated with targeted studies that introduce additional variables.
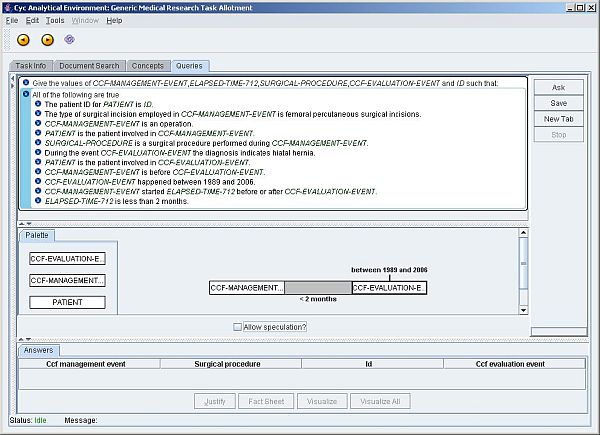
Figure 4: Fully specified query in the Semantic Web Medical Expert System (a larger version of the screen dump is also available)
Use of RDF vocabulary to (explicitly) describe structural constraints facilitates the automated generation of much of the application logic associated with management of instance data over the lifetime of the repository. For example, in addition to XML->RDF transforms and the XML schemas, the XForms infrastructure used for data entry is generated in such a fashion.
Finally, exporting the domain as OWL facilitates inexpensive, highly-automated, integration with external systems that have native capabilities for more expressive languages. The mapping to the Cyc general consensus ontology was driven completely by semantics of the domain rather than how the data were physically constituted. The latter is typically much more emphasized with integration efforts across database management systems that use less expressive data formats.
So, the key Semantic Web advantages are:
© Copyright 2007, Cleveland Clinic